Table 2.
Structure and mechanism of hRR inhibitors.
| Structure | Name | Cellular Mechanism | RR Subunit Targeted | Mechanism of RR Inhibition |
|---|---|---|---|---|
|
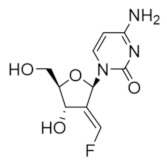
Tezacitabine | Interferes with DNA synthesis and repair mechanisms | hRRM1 | Irreversibly modifies the C-site |
|
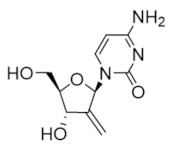
2′-deoxy-2′-methylenecytidine (DMDC) | Interferes with DNA synthesis and repair mechanisms | hRRM1 | Inhibits the C-site |
|
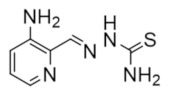
Triapine | Metal chelation | hRRM2 | Chelates FeIII from the radical center |
|
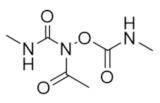
Caracemide | Radical Scavenger | hRRM1 | Quenches the radical center |
|
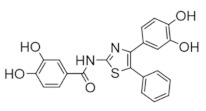
COH29 | Interferes with DNA synthesis and repair mechanisms | hRRM2 | Binds hRRM2 near the C-terminal tail to prevent association with hRRM1 |
|
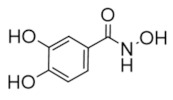
Didox | Radical Scavenger | hRRM2 | Chelates FeIII from the radical center |
|
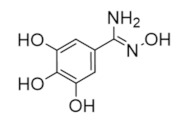
Trimidox | Radical Scavenger | hRRM2 | Chelates FeIII from the radical center |
|
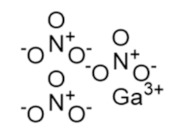
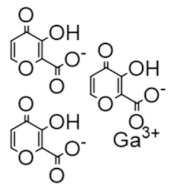
Gallium nitrate | FeIII mimic | hRRM2 | Interferes with FeIII radical center |
|
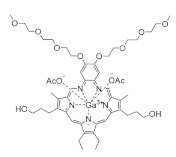
Gallium maltolate | FeIII mimic | hRRM2 | Interferes with FeIII radical center |
|
Motexafin gadolinium | FeIII mimic | hRRM2 | Interferes with FeIII radical center |
| GTI-2040 | antisense ologonucleotide | hRRM2 | Inhibits hRRM2 | |
| GTI-2501 | antisense ologonucleotide | hRRM2 | Inhibits hRRM2 | |
| CALAA-01 | siRNA-containing nanoparticle | hRRM2 | Inhibits hRRM2 |
