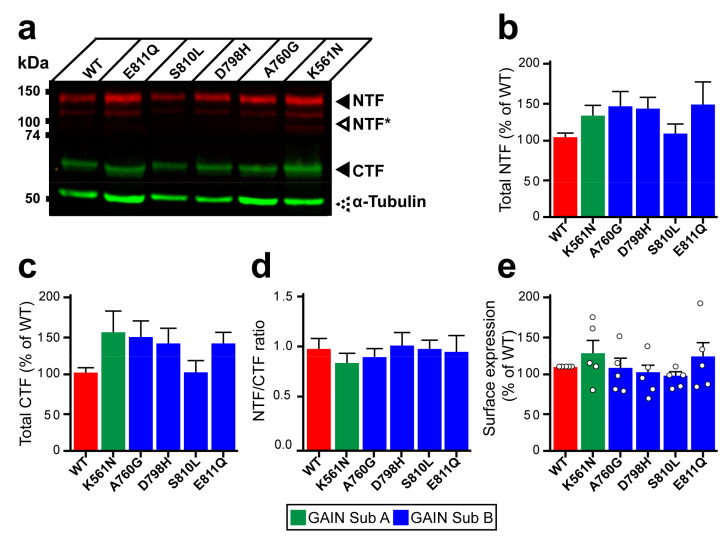
Figure 3.
Expression and autoproteolysis of Lphn3 are unaltered by the presence of cancer-related GAIN domain mutations. (a) Western blot analysis by immunofluorescent detection of total proteins from HEK293 cells transfected with cancer-related GAIN domain mutations. NTF was detected with a rabbit anti-Flag antibody and CTF with a mouse anti-HA antibody, both distinctly labeled with corresponding secondary fluorescent antibodies. NTF* represent fragments resulting from unknown posttranslational modifications. Immunodetection of α-tubulin was used as a protein loading control. (b,c) Quantification of normalized fluorescent immunodetection data obtained in (a) for NTF and CTF, respectively. (d) Cleavage efficiency quantification expressed as NTF/CTF ratios. Data from b-d are represented as the mean values obtained from at least four independent experiments with error bars representing S.E.M. (n = 4). (e) Detection of cell surface expression assay (DECS) for Flag-tagged Lphn3 and cancer-related GAIN domain mutations using a colorimetric reaction quantifying the acid-stopped conversion of Horseradish-peroxidase substrate, which is detected at a 450 nm absorbance wavelength. Subdomain A (GAIN Sub A) and B (GAIN Sub B) of GAIN domain. Statistical analysis was performed using one-way ANOVA. Each white circle symbol represents the mean value of four replicates obtained from one independent experiment. Data were normalized to Lphn3-WT and are represented as the mean values obtained from at least five independent experiments of four replicates, each with error bars representing S.E.M. (n = 5).