Figure 1. Schematic diagram showing structure and design of a typical zinc finger nuclease.
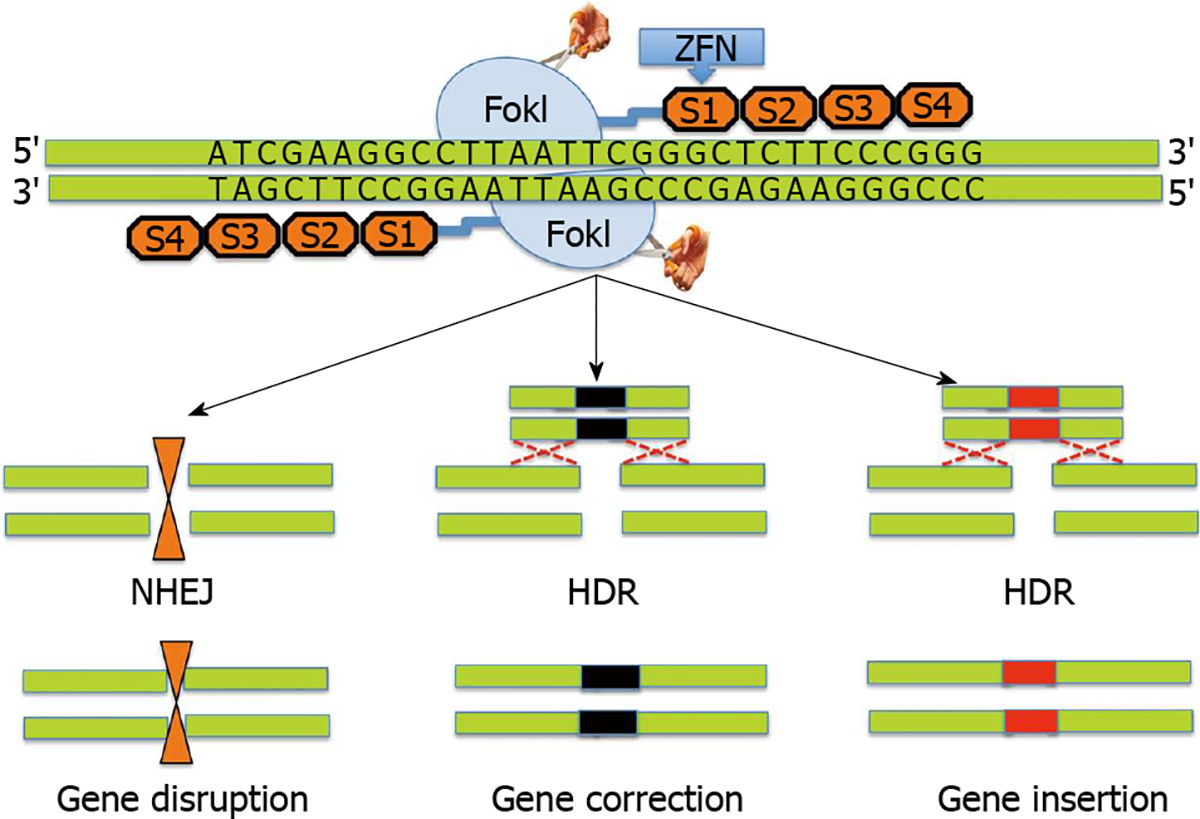
Zinc finger nucleases (ZFNs) use a modular array of 3–6 ZFNs (4 shown) specifically designed to bind to the target DNA together with the FokI cleavage domain. The FokI cleavage domains can be engineered to function as heterodimers or homodimers to achieve desired cleavage specificity. ZFNs typically recognize 24–36 bp unique sequence within the genome to achieve target specificity. ZFN mediated cleavage of the target leads to double strand breaks, which in turn induces either non-homologous end joining pathway (NHEJ) or homology directed repair (HDR) processes. NHEJ leads to gene disruption due to small insertions or deletions (indels) while HDR leads to gene correction.
