Figure 3. Clustered Regularly Interspaced Short Palindromic Repeat/Clustered Regularly Interspaced Short Palindromic Repeat Associated Systems.
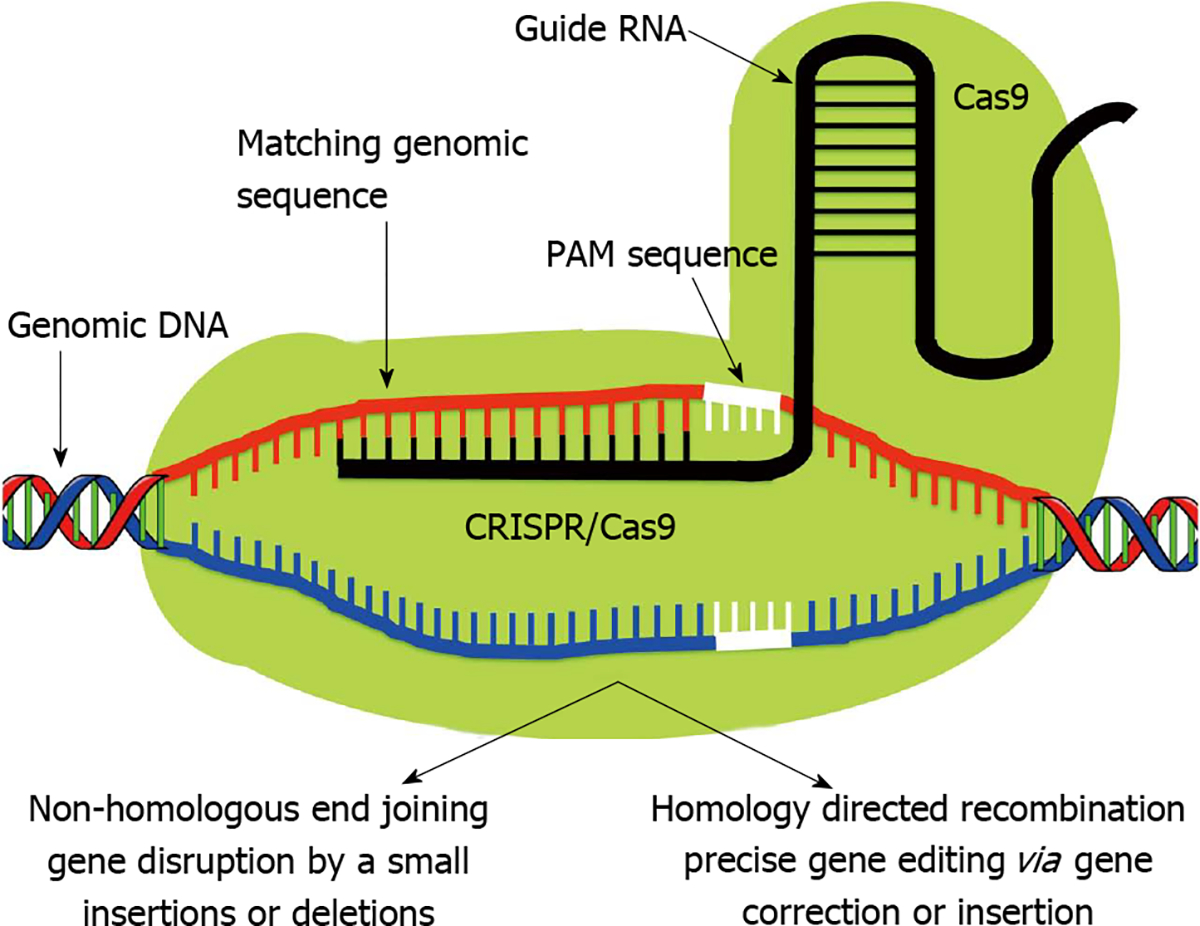
In contrast to Like zinc finger nucleases and transcription activator-like effector nucleases, Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)-associated protein (Cas9) monomer possess innate nuclease activity which catalyzes double strand breaks leading to random knockout phenotypes via non-homologous end joining pathway. Therefore Cas9 requires a single guide RNA (sgRNA) to recognize its target site. The sgRNA is composed of two separately expressed RNAs including a CRISPR RNA (crRNA) and a trans-activating crRNA (tracrRNA), which are processed by endogenous bacterial machinery to yield the mature gRNA. The current CRISPR/Cas9 system employs a single chimeric sgRNA, which is a fusion of crRNA and tracrRNA. Currently used sgRNA typically contains a 17–20 nucleotide long variable region, which is complementary to the genomic target sequence. A short region immediately 3’ to the target sequence known as protospacer adjacent motif has NGG sequence which is a major specificity determinant of Cas9. PAM: Protospacer-adjacent motif.
