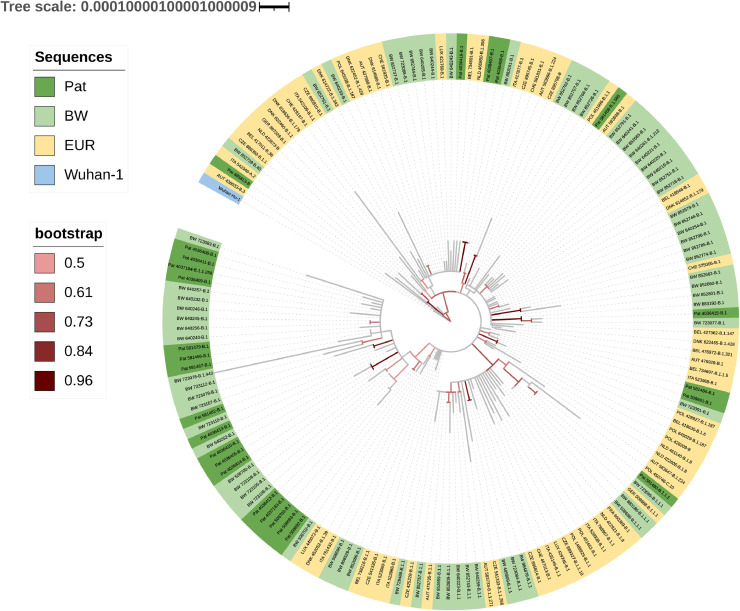
Fig. 4.
Maximum likelihood phylogenetic tree of SARS-CoV-2.
The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Pat = patient-derived sequences (n = 26). BW = Baden-Wuerttemberg, Germany (n = 69). EUR = Germany and it's neighboring countries (n = 67). Wuhan-1: reference root sequence. 1000 bootstrapping iterations were used. Branches with bootstrapping values above 50% are colored.