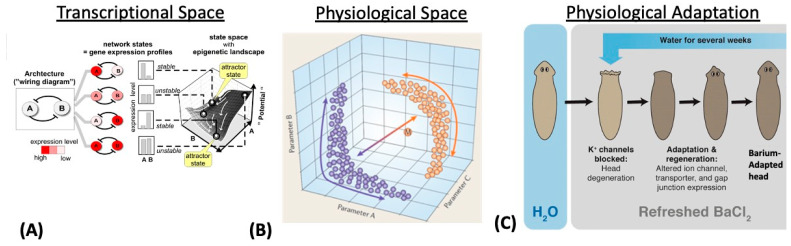
Figure 2.
Diverse spaces within which living systems navigate. Transcriptional space is the space of possible gene expression patterns, taken with permission from [59]. Examples of a transcriptional state space of a two-gene network (mutual inhibition of genes (A,B)) and the associated epigenetic landscape in the two-dimensional state space are shown. The dynamical state of the network maps to a point in the space; changes in gene activity represent walks in the space. (B) Physiological space is the space of possible physiological states, simplified to just three parameters such as intracellular ion concentrations, taken with permission from [70]. Individual cells occupy regions of the space and can move between states by opening and closing specific ion channels. The functional state (region of the space) is a function of all of the parameters and large-scale variables, such as Vmem (resting potential), which refer to numerous microstates composed of individual ion levels. (C) Navigating spaces to thrive despite novel stressors, taken with permission from [71]. Planaria exposed to barium chloride experience head deprogression because barium is a blocker of potassium channels, making it impossible for the neural tissues in the head to maintain a normal physiology. The flatworms soon regenerate a new head which is barium-insensitive. Transcriptomic analysis showed only a handful of genes whose expression was altered in the barium-adapted heads. Because barium is not something planaria encounter in the wild, this example shows the ability of the cells to navigate transcriptional space to identify a set of genes that enable them to resolve a novel physiological stressor. The mechanism by which they rapidly determine which of many thousands of genes should be up- or downregulated in this scenario is not understood. (The cells do not turn over fast enough to allow a hill-climbing search, for example.)