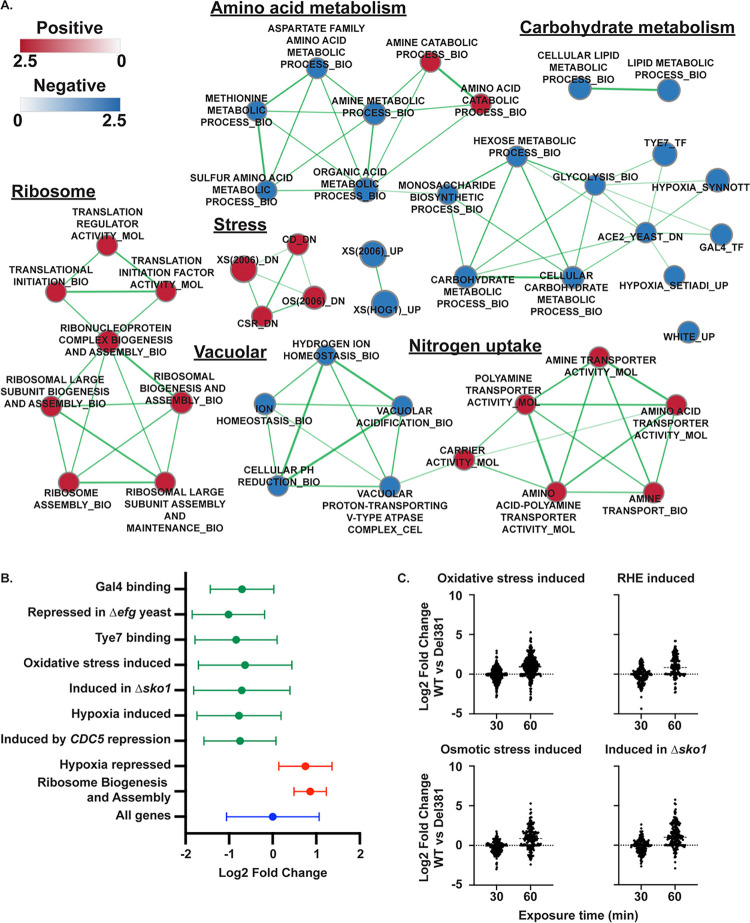
Fig 11. Cells containing derepressed TOR1-Del381 upregulated ribosome biogenesis and translation related genes but decreased expression of carbon metabolism- and stress response genes.
A. Enrichment map showing the major categories of genes identified by Gene Set Enrichment Analysis (GSEA) in the transcript profile of tor1/tetO-TOR1-Del381 cells (JKC1441) grown in YPD without doxycycline compared to the heterozygote tor1/TOR1 (JKC1347). Nodes represent gene sets (FDR q <0.1) where color intensity reflects the Normalized Enrichment Score (NES) and node size reflects the number of enriched genes. Width of connecting lines (edges) indicates the degree of overlap between categories. B. Gene set enrichment analysis was used to identify major categories of genes significantly regulated (adjusted P value < 0.001) in tor1/tetO-TOR1-Del381 cells (JKC1441) compared to the tor1/TOR1 control (JKC1347). Graph shows the fold change (Log2) of genes in each indicated category in Del381 cells relative to tor1/TOR1 cells. Full results in S1 File and S2 File. C. Specific gene categories were altered in cells lacking N-terminal HEAT repeats during superoxide stress. Comparison of gene expression in wild type (WT, SC5314) relative to tor1/tetO-TOR1-Del381 (JKC1441) in the presence of plumbagin (10 μM) after 30 and 60 minutes. Gene categories shown were significantly represented (Q < 0.001) among genes exhibiting increased expression in the wild type relative to tor1/tetO-TOR1-Del381 cells following 60 minutes incubation in plumbagin. Gene categories are those identified by Enjalbert et al. (oxidative and osmotic stress;[67]), Rauceo et al. (Δsko1; [112]) and Spiering et al. (Reconstituted human epithelium induced; [113]).