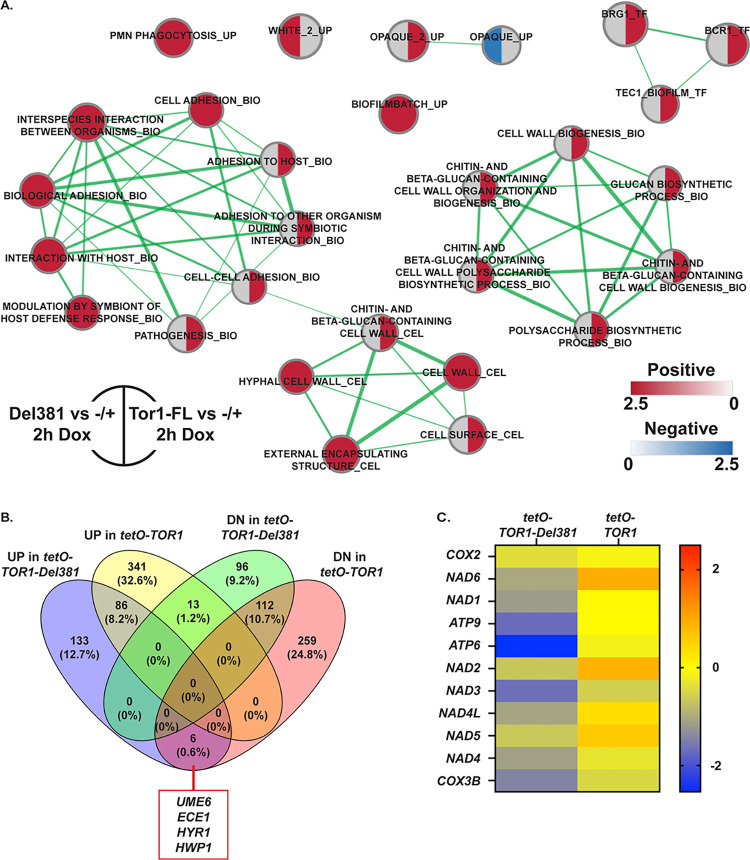
Fig 12. Comparison of gene set expression changes during repression of TOR1 and TOR1-Del381 in FL and Del381 cells exposed to doxycycline.
A. Enrichment map showing the major categories of genes identified by Gene Set Enrichment Analysis (GSEA) in the transcript profiles of tor1/tetO-TOR1-Del381 cells (JKC1441, left side) and tor1/tetO-TOR1 cells (JKC1549, right side) compared to heterozygous tor1/TOR1 cells (-/+, JKC1347) following tetO repression in YPD with doxycycline for 2 h. Nodes represent gene sets (FDR q <0.1) where color intensity reflects the Normalized Enrichment Score (NES) and node size reflects the number of enriched genes. Width of connecting lines (edges) indicates the degree of overlap between categories. Grey indicates the category was not significant under the experimental condition. B. Comparison of gene expression changes (≥2-fold up and down) in tor1/tetO-TOR1-Del381 (JKC1441) and tor1/tetO-TOR1 (JKC1549) cells exposed to doxycycline for 8 h. Six genes induced in tor1/tetO-TOR1-Del381 cells were suppressed in tor1/tetO-TOR1 cells, including UME6, ECE1, HYR1 and HWP1. C. Heatmap showing expression of genes that encode Complex I of the mitochondrial electron transport chain in tor1/tetO-TOR1-Del381 (JKC1441) relative to the heterozygote tor1/TOR1 (column 1) and in tor1/tetO-TOR1 (JKC1549) relative to the heterozygote tor1/TOR1 (column 2). Expression values are Log2 values relative to tor1/TOR1 (JKC1347).