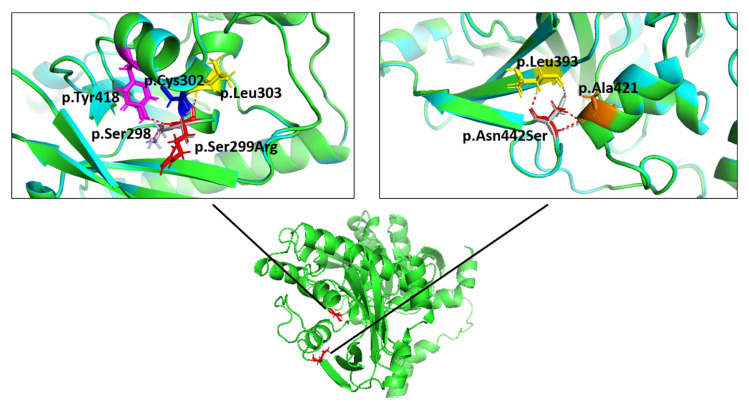
Figure 5.
Molecular modelling of the FKTN variants p.Ser299Arg (left panel) and p.Asn442Ser (right panel), with structures calculated using ColabFold [23]. The influence of the variants on the molecular structure was analyzed using PyMOL Molecular Graphics System (version 2.5.2, Schrodinger LLC, New York, NY, USA). The protein backbone is shown in green for the wildtype and in cyan for the respective mutant. No major structure change was observed in the alignment between the wildtype and mutant proteins, as evidenced by the protein backbone overlay. The polar contacts were calculated using PyMOL and are shown as grey or red dashed lines for the wildtype or mutant proteins, respectively. The serine at position 299 (left panel, p.Ser299, grey) has polar contacts to p.Ser298 (purple), p.Cys302 (blue), and p.Leu303 (yellow). In the p.Ser299Arg mutant (left panel, red), an additional polar contact to Tyr418 (magenta) was observed. The asparagine and serine at position 442 (right panel, grey: p.Asn442, red: p.Ser442) had polar contacts to p.Ala421 (orange) and p.Leu393 (yellow).