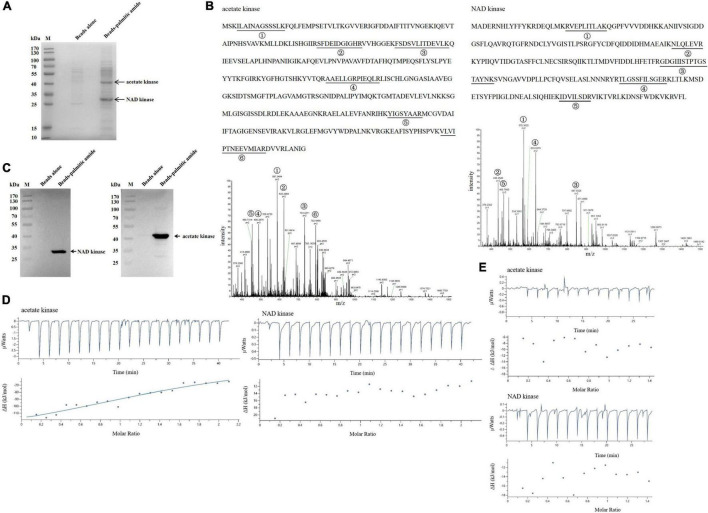
FIGURE 4.
The proteins interacted with palmitic amide. (A) The proteins bound to palmitic amide. Geobacillus sp. E263 was lysed by ultrasonication and the proteins in supernatant were incubated with palmitic amide-coupled NHS-activated beads. The proteins were separated by SDS-PAGE with Coomassie brilliant blue staining. NHS- activated beads alone was used as a control. The arrows represented the differential proteins. M, protein marker. (B) Identification of the proteins bound to palmitic amide by mass spectrometry. The proteins were identified to be acetate kinase and nicotinamideadenine dinucleotide (NAD) kinase, respectively. The matched peptides were indicated with underlines. (C) Western blot analysis of the proteins interacted with palmitic amide. The proteins extracted from Geobacillus sp. E263 were incubated with palmitic amide-coupled beads, and then subjected to Western blot using NAD kinase-specific or acetate kinase-specific antibody. The target protein was indicated with an arrow. M, protein marker. (D) Interaction between palmitic amide and acetate kinase or NAD kinase. The purified recombinant acetate kinase (20 μM) or NAD kinase (20 μM) was incubated with palmitic amide (200 μM) to conduct isothermal titration calorimetry (ITC). (E) Evaluation of the binding of palmitic acid to acetate kinase or NAD kinase by ITC. Palmitic acid (200 μM) was incubated with the purified recombinant acetate kinase (20 μM) or NAD kinase (20 μM) to perform ITC.