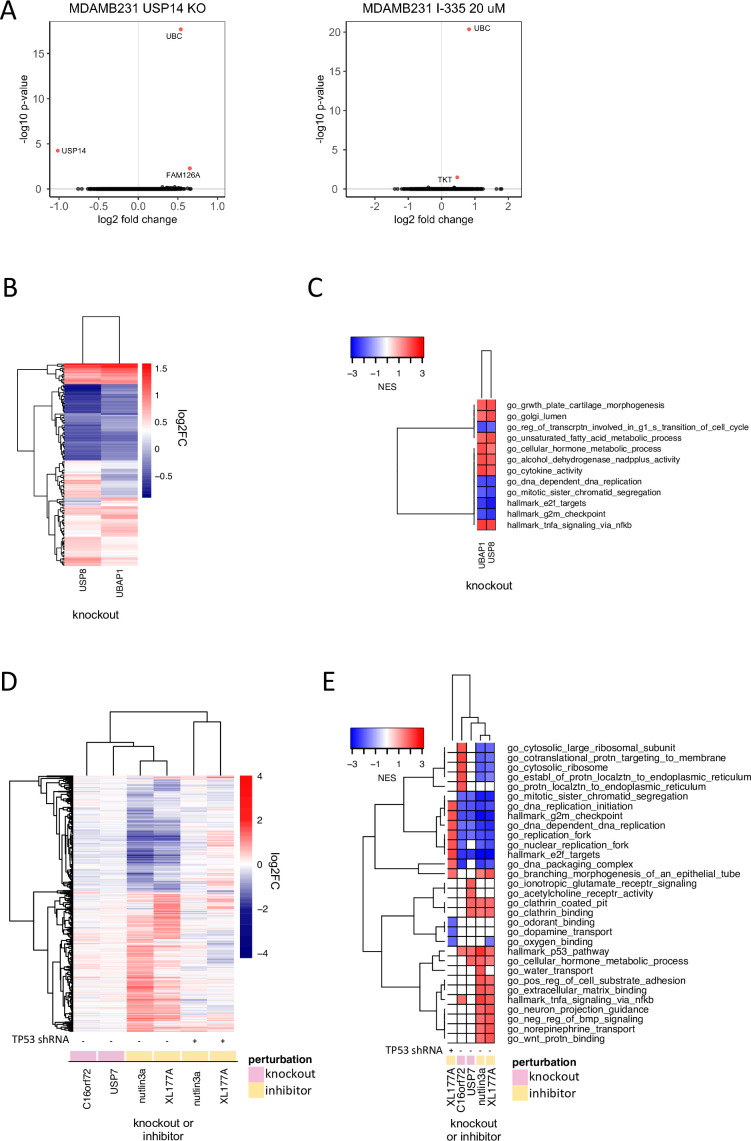
Figure 4. Discriminating the functions of well-studied DUBs.
(A) Changes in gene expression (log2 fold change vs. log 10 adjusted p-value, adjusted p-value <0.05 colored red) in MDAMB231 cells 96 hr following USP14 knockout by CRISPR-Cas9 (left) or 24 hr after treatment with the USP14 inhibitor I-335 at 20 µM (right). (B) Hierarchical clustering of significantly differentially expressed genes (adjusted p-value <0.05) 96 hr following knockout of USP8 or UBAP1 in MDAMB231 cells. (C) Gene sets significantly (FDR <0.05) enriched in MDAMB231 cells 96 hr after knockout of USP8 or UBAP1. The top five upregulated and top five downregulated gene sets for each condition are shown. (D) Hierarchical clustering of significantly differentially expressed genes (adjusted p-value <0.05) 96 hr after knockout of USP7 or C16orf72 in wild-type MCF7 cells or following a 24 hr treatment with 5 µM nutlin3a or 1 µM XL-177A in wildtype or p53 knockdown MCF7 cells. (E) Gene sets significantly enriched (FDR <0.05) for the conditions shown in (D). The top five up- and down-regulated gene sets for each condition are shown.