Figure 7.
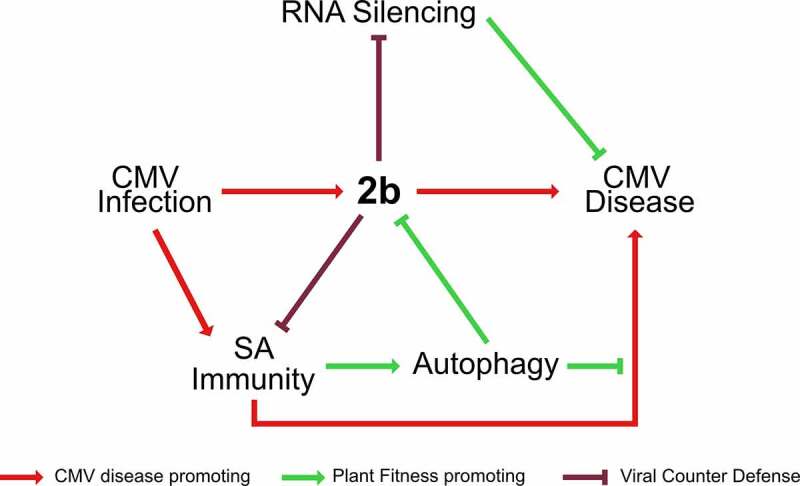
Model for the interplay between autophagy, SA, RNA silencing and 2b in CMV disease. The 2b protein is the major pathogenicity determinant in CMV infection. SA activates autophagy upon CMV infection and mediates 2b degradation. By limiting 2b levels, autophagy relaxes 2b mediated suppression of antiviral RNA silencing and virulence. Vice versa, 2b itself has the capacity to restrict both SA-dependent autophagy and plant growth reduction, predicting complex interactions between SA, autophagy and 2b in CMV disease. Furthermore, SA, autophagy and antiviral RNA silencing pathways all suppress virus accumulation, and the lack of additivity between the pathways suggests the possibility that they interact in the process, potentially through 2b degradation and SA-dependent autophagy. Taking SA-driven disease development under autophagy deficiency together with the prominent synergism of autophagy and RNA-silencing in disease attenuation, we propose that autophagy and RNA silencing-based plant health is not quantitatively coupled to virus accumulation and that the pathways rather operate in a parallel manner to promote survival of infected plants. Virulence evolution and trade-offs are complicated for pathogens that utilize both vertical and horizontal transmission. We consider that CMV has adapted to and benefits from these potential antiviral pathways. Thus, the interplay between the viral 2b protein, SA, plant autophagy and RNA silencing pathways determines the delicate balance between virus accumulation, transmission and plant fitness in CMV disease.
