Figure 3.
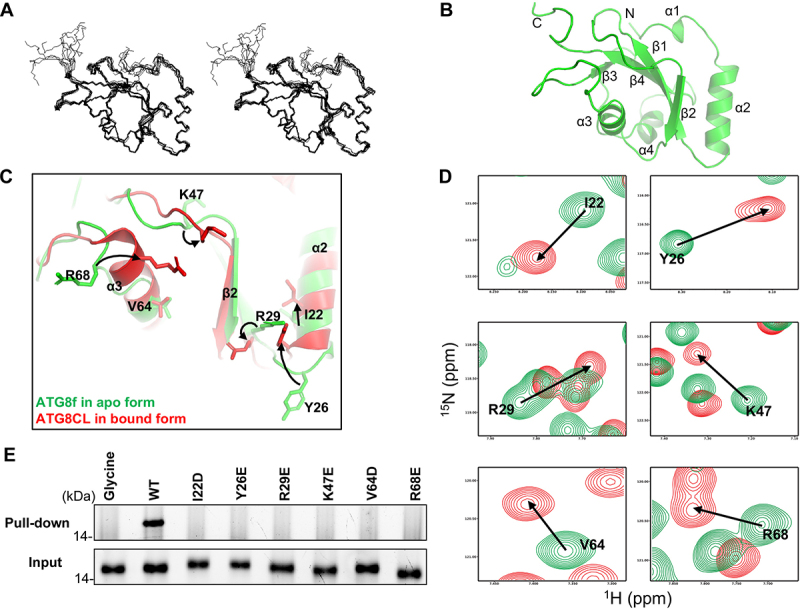
Binding of the NBR1 AIM peptide induces conformational changes around the ligand binding sites in ATG8f. (A) Solution structure of ATG8f. Stereodiagram of an ensemble of the 10 best structures showing the best-fit superposition of the backbone atoms. (B) Ribbon representation of the representative ATG8f structure. N- and C- termini and secondary structure elements were labeled. (C) Structure of Arabidopsis ATG8f in apo form (green) was compared to that of Irish potato ATG8CL in complex with the AIM peptide (red) of PexRD54 (PDB code: 5L83). Upon binding of the cognate AIM peptide, I22, Y26, R29, K47, R68 with conformation changes were evident in regions around the ligand binding sites of ATG8. (D) 1H-15N correlation spectra of ATG8f in the absence (green contours) and in the presence (red contours) of the NBR1 AIM peptide (657–667 aa: GVSEWDPILEE) were compared. Significant chemical shift perturbations were found for residues (I22, Y26, R29, K47, V64, R68) around the ligand binding sites of ATG8f, suggesting binding of the NBR1 AIM peptide induced changes in the chemical environment around these residues. (E) The role of residues with large chemical shift perturbations was tested by mutagenesis and affinity-isolation assay. 1 μM wild-type or variants of ATG8f was mixed with NHS-activated Sepharose resins coupled with the NBR1 AIM peptide, and the bound proteins were stained by Instant-Bands and analyzed by SDS-PAGE. Our results showed that all substitutions weaken the interaction between ATG8f and the NBR1 AIM peptide, suggesting these residues are important in the interaction. Wild-type ATG8f loaded to the NHS-activated Sepharose coupled with glycine was included as a negative control.
