Figure 6.
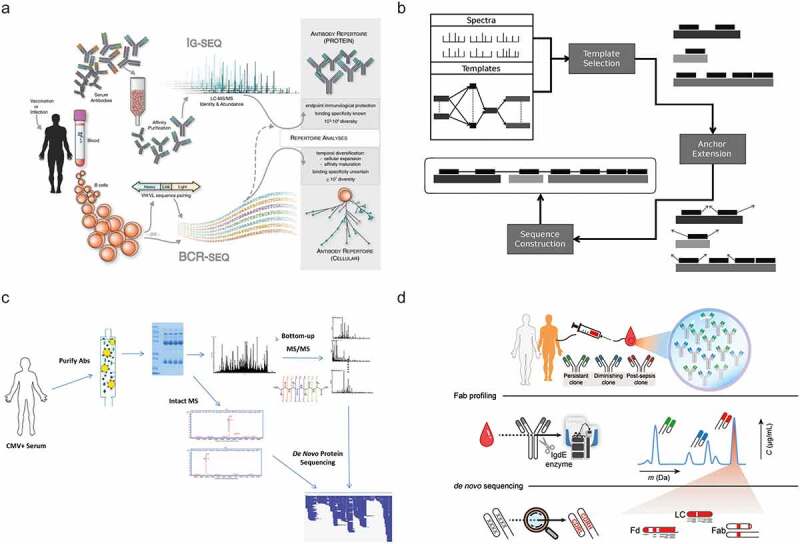
Selected recent approaches aiming toward MS-based de novo sequencing of serum antibodies. (a) In Ig-seq.90 a personalized database generated by BCR sequences is used to identify specific clones, using tryptic peptides covering the CDR3 region. Figure adapted from Lavinder et al.90 (b) GenoMS47 uses genomic data to generate template sequences. The specific construction of the templates can be defined by the user from either whole genome sequencing or BCR sequencing data. Figure adapted from Castellana et al.47 (c) PolyExtend32 helped to analyze a polyclonal mixture of antigen-specific purified antibodies measured by BU MS and intact mass measurements. Using a user-assisted algorithm, these data from different MS modalities were combined to sequence the most abundant clones. Figure adapted from Guthals et al.32 (d) Fab profiling31 measures and quantifies intact masses of Fabs to provide a view of the IgG1 clonal repertoire, enabling to quantify and monitor individual clones. Abundant serum clones are identified by using BU and MD MS data iteratively to generate full IgG de novo sequences. Figure adapted from Bondt et al.31
