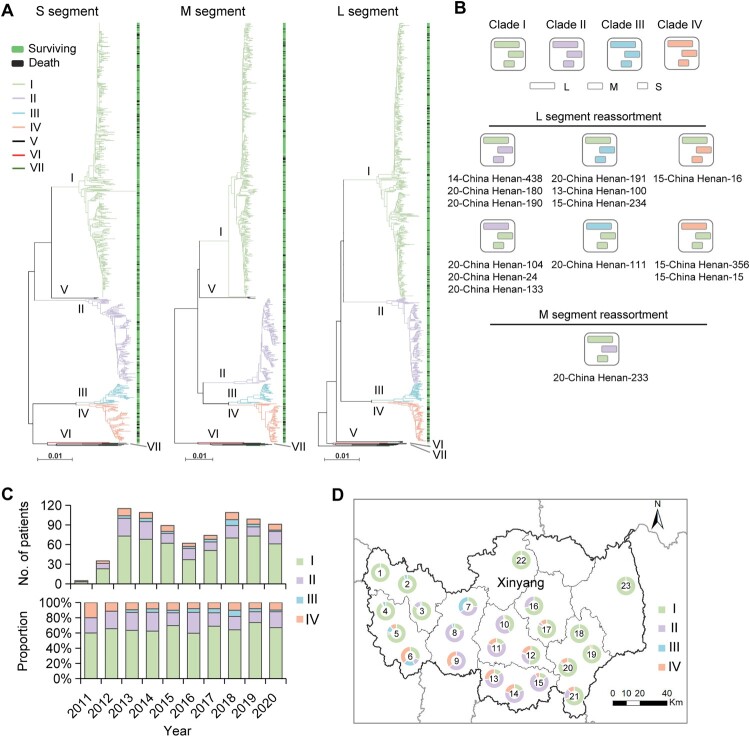
Figure 1.
Phylogenetic characterization of SFTSV S, M, and L segments sequenced from human patients. Maximum-likelihood trees for SFTSV S, M, and L genome segments (A). Seven viral clades indicated by coloured branch were identified from patients with SFTS in this study. The black branch indicates sequences downloaded from the GenBank. The outcome (surviving or death) of the patient was shown on the right side of the phylogenetic trees. Scale bars indicate a number of substitutions per site. Graphic representation of reassortment events in SFTSV genome sequences obtained from patients (B). The number beneath the reassortment graphics indicates the name of the virus isolates with detailed information in Appendix Table 2. The number and proportion of four common viral clades obtained from patients with SFTS in each studied year (C). Geographic distribution of four common viral clades from patients with SFTS (n = 712) residing in Xinyang City (D). The number in the middle of the circle indicates the region number where the patients reside, with detailed data shown in Table S7.