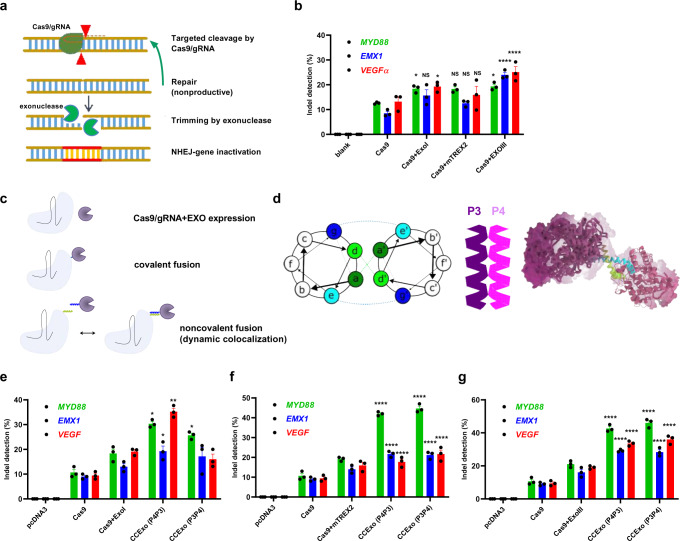
Fig. 1. Tethering of Cas9 to exonucleases via coiled-coil dimer peptides for the enhanced efficiency of genome editing.
Schematic presentation of exonucleases on excessive DNA trimming for enhanced gene inactivation (a). HEK293 cells (2*105 cells/ml) were co-transfected with plasmids encoding Cas9, gRNA and an exonuclease (human ExoI, mouse TREX2 or E.coli ExoIII) (600 ng of each component). 48 h later genomic DNA was isolated and T7E1 assay for indel detection was performed. Data present three separate experiments (n = 3). *P < 0.05, ****P < 0.0001. All P values are from ordinary one-way ANOVA followed by Tukey’s multiple comparisons test to Cas9 values. Data are presented as mean values ± SEM as appropriate (b). Different approaches for achieving increased gene editing using Cas9 and exonucleases (c). Schematic presentation of P3 and P4 parallel coiled-coil dimer interactions tethering Cas9 and exonuclease with green and blue letters presenting hydrophobic and charged interacting amino acid residues, respectively (d). HEK293 cells (2*105 cells/ml) were transfected with plasmids encoding gRNA and CCExo (Cas9-P3 or -P4, P3- or P4-ExoI, P3 or P4-mTREX2 or P3- or P4-ExoIII) (600 ng of each component). 48 h later genomic DNA was isolated and T7E1 assay was performed to assess indel frequency. Indel frequencies were compared between editing with Cas9 alone, co-expression of Cas9 with an exonuclease or coiled-coil based coupling of Cas9 to an exonuclease, wherein different fusion variants of P3 and P4 coils were tested (P4P3 denotes transfection with Cas9-P4 coexpressed with P3-Exo- and P3P4 denotes Cas9-P3 coexpressed with P4-Exo). Indel frequencies were tested for human EXOI (e), mouse TREX2 (f) and E.coli ExoIII (g). Data present results from three separate experiments (n = 3). *P < 0.05, **< 0.01, ***< 0.001 and ****P < 0.0001. All P values are from ordinary one-way ANOVA followed by Tukey’s multiple comparisons test compared to Cas9 values. Data are presented as mean values ± SEM as appropriate.