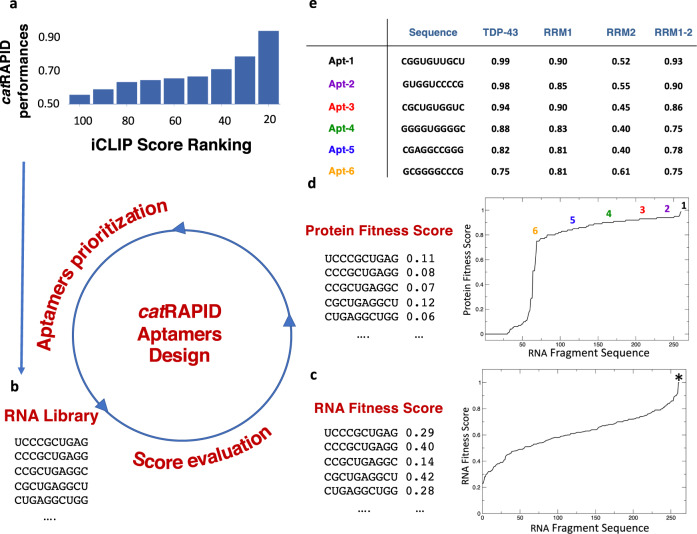
Fig. 1. In silico design of RNA aptamers.
Sketch of the computational pipeline. a TDP-43 interaction propensity computed with catRAPID is used to discriminate between interacting (high iCLIP score) and non-interacting (low iCLIP scores) transcriptomic regions. The iCLIP scores were ranked from low (100 transcriptomic regions with the highest iCLIP score vs 100 transcriptomic regions with the lowest iCLIP score) to high (20 transcriptomic regions with the highest iCLIP score vs 20 transcriptomic regions with the lowest iCLIP score). catRAPID performances increase with the strength of the iCLIP score, indicating that the algorithm can accurately identify strong-signal interactions. b Using the 30 top-interacting transcriptomic regions we generated a list of fragments using a sliding window of 10 nucleotides moved from the 5′ to 3′ of each sequence. c The RNA Fitness score was employed to measure the strength of catRAPID interaction propensities upon mutation of the RNA fragment sequence. Examples of RNA fragments and their relative RNA Fitness scores are shown on the left panel. On the right panel, the RNA Fitness scores of all the RNA fragments generated are given (candidate aptamers were marked with a star). d The Protein Fitness score measures the strength of interaction propensity of each RNA sequence for TDP-43 in comparison with a pool of protein sequences with the same length and amino acid composition. Examples of RNA fragments and their Protein Fitness scores are shown on the left panel. On the right panel, the RNA Fitness scores of the RNA fragments generated are given (candidate aptamers were marked with different numbers and colors). e Summary of Protein Fitness scores for TDP-43 full-length, RRM1, RRM2, RRM1-2 sequences (the colors match the Protein Fitness reported in panel d).