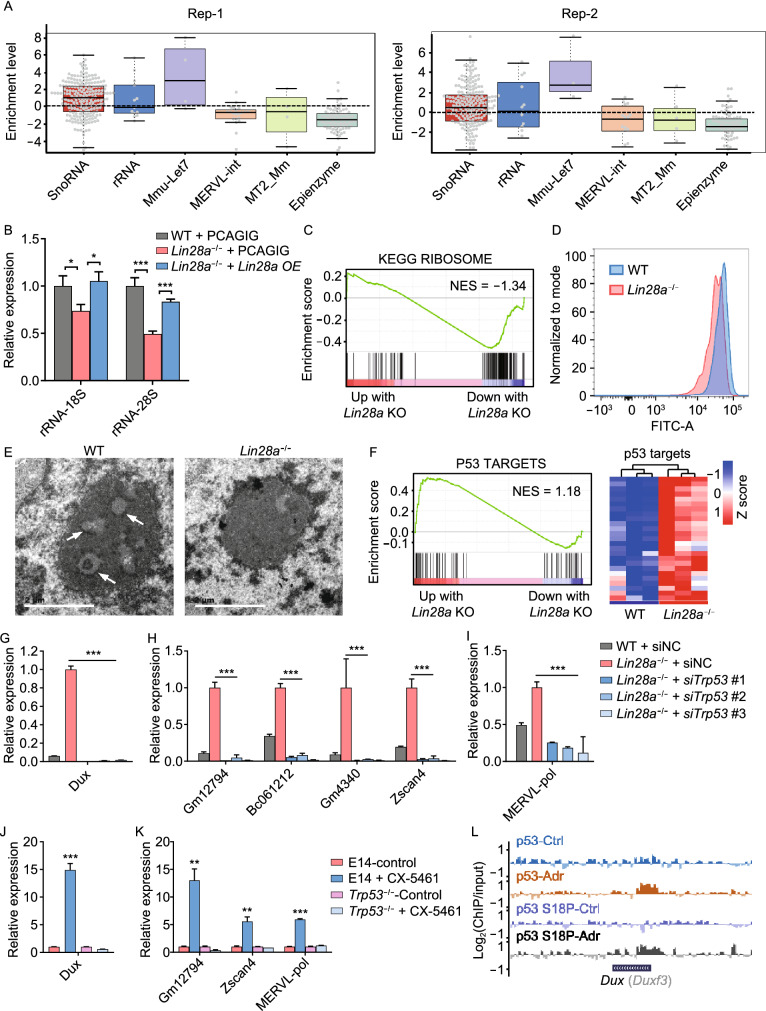
Figure 4.
LIN28 binds to snoRNA and rRNA, and its loss leads to reduced rRNA biogenesis and ribosomal stress. (A) LIN28A CLIP-seq enrichment analysis. ERVL, MERVL-int and MT2_Mm classes were not enriched in LIN28A binding targets compared with other genes. (B) qRT-PCR showing the expression of rRNA in wild-type and Lin28a knockout ES cells. PCAGIG: empty vector. OE: overexpression. *P < 0.05, ** P < 0.01, *** P < 0.001, two-way ANOVA, n = 3, error bar: standard error of the mean. (C) GSEA analysis showing collective changes in the Ribosome gene set in Lin28a knockout ES cells compared with wild-type cells. NES: normalized enrichment score. (D) Flow cytometry showing OP-puromycin (OP-puro) labeling of wild-type and Lin28a knockout ES cells. (E) Electro-microscopy showing nuclear morphology of wild-type and Lin28a knockout ES cells. Scale bar, 2 μm. (F) Left panel: GSEA analysis showing collective changes in the p53 signaling pathway gene set. Right panel: up-regulated p53 target genes in Lin28a knockout ES cells compared with wild-type cells. (G–I) qRT-PCR showing Dux (G), 2C-related genes (H) or MERVL (I) expression in wild-type and Lin28a knockout ES cells treated with scramble negative control, or Trp53 siRNAs. (J and K) qRT-PCR showing the expression of Dux (J) or 2C-related genes (K) in ES cells treated with 4 μmol/L CX-5461 for 12 h or equal water (control), or in Trp53 knockout ES cells treated with the same conditions. (L) UCSC Genome Browser view of ChIP-seq results using antibodies of p53 and phosphorylated p53 S18P for Dux gene locus. *P < 0.05, **P < 0.01, ***P < 0.001, two-way ANOVA, n = 3, error bar: standard error of the mean