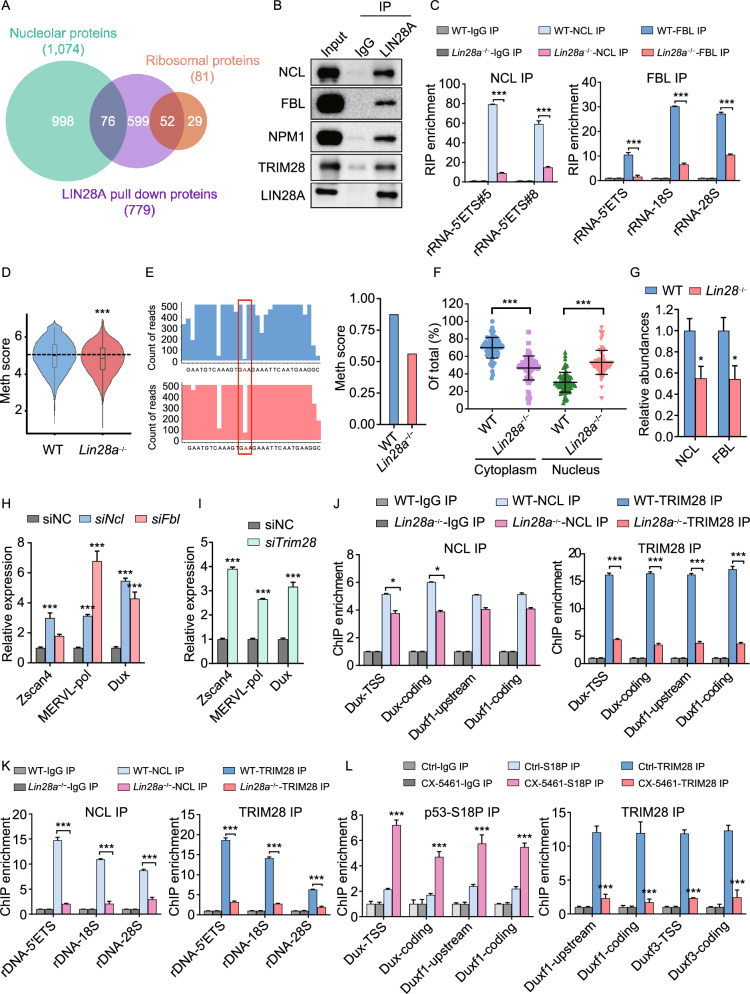
Figure 5.
LIN28A is associated with nucleolar proteins and mediates rRNA biogenesis and 2C gene repression through the NCL/TRIM28 complex. (A) Venn diagrams showing the overlap between proteins pulled down by LIN28A and nucleolar proteins or ribosomal proteins. (B) Validation of the interaction between the endogenous LIN28A and nucleolar or nuclear proteins by Co-IP followed by Western blot analysis in ES cells. IgG was used as a negative control for the IP. (C) RIP-qPCR assays for NCL and FBL in wild-type and Lin28a knockout ES cells. *P < 0.05, **P < 0.01, ***P < 0.001, two-way ANOVA, n = 3, error bar: standard error of the mean. (D) Quantification of 2’-O-Me by RiboMethseq for all bases in 28s, 18s and 5s rRNAs in wild-type and Lin28a knockout ES cells. MethScore was calculated using the method mentioned in (Birkedal et al., 2015; Marchand et al., 2016). ***P < 0.001. Wilcox signed rank test. (E) Left panel: RiboMeth-seq showing profiles of mapped sequencing reads in a range of a snoRNA targeting region on the 28s rRNA (from 3,366 bp to 3,397 bp). Red highlighted A is a predicted methylation site (from SnoRNA Orthological Gene Database). Right panel, calculated RiboMeth-Seq score to indicate the level of the 2’-O-methylation at the predicted site of A (Birkedal et al., 2015). (F) Proteomics analysis showing the nucleus and cytoplasm proportion of ribosomal proteins in wild-type and Lin28a knockout ES cells. ***P < 0.001, t-test, n = 3, error bar: standard error of the mean. (G) Proteomics analysis showing the relative abundances of indicated proteins in wild-type and Lin28 knockout cells (averaged proteomics data from Lin28a knockout ES cells and Lin28a/b knockout iPS cells). Error bar: standard error of the mean. n = 2 independent proteomics experiments. (H and I) qRT-PCR showing the expression of 2C-related genes in ES cells treated with scramble negative control or the indicated siRNAs. (J and K) NCL and TRIM28 ChIP-qPCR analysis at the Dux (J) and rRNA (K) loci in wild-type and Lin28a knockout ES cells. (L) p53 and TRIM28 ChIP-qPCR analysis at the Dux loci in untreated and CX-5461-treated ES cells