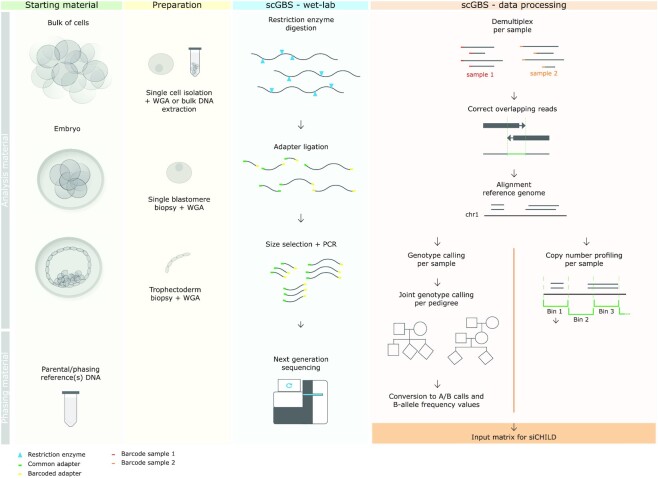
Figure 1.
scGBS can be performed on DNA extracted from multiple cells, e.g. a cell line, or single cells, e.g. from embryos samples. Parental DNA and DNA from family members, such as grandparents or siblings, with respect to the analysis sample(s), is used for haplotyping. Preparation for scGBS consists of isolation of a single cell or multiple cells followed by a whole genome amplification (WGA) by multiple displacement amplification (MDA). Subsequently, the amplified genomic DNA is digested by a restriction enzyme followed by adapter ligation, size selection and a PCR. Multiplexing of samples is performed after adapter ligation. Processing of scGBS data consists of demultiplexing raw sequencing reads per sample, correction of overlapping reads by FLASH (40) and alignment of the corrected reads to the reference genome with BWA MEM (42). Next, GATK Haplotypecaller is applied to each sample separately and GATK GenotypeGVCFS allows for joint genotype calling per pedigree with samples to analyze combined with parental and phasing reference samples (43). A customized conversion is applied to obtain the desired input for siCHILD analysis such as A/B calling and B-allele frequency (BAF). In parallel, after the alignment step a copy-number profiling is performed per sample with QDNAseq (46). Finally, discrete genotypes, BAF and logR values are combined into one input matrix for siCHILD analysis.