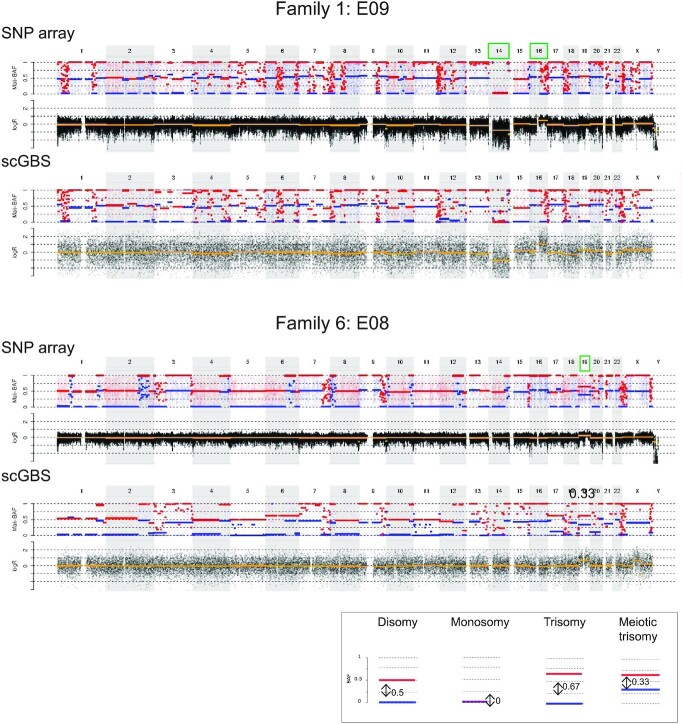
Figure 5.
An example of the genome-wide comparison between scGBS- and SNP-array-based copy-number profiling is shown for two embryo biopsies from two families. Only the genome-wide maternal haplarithm (Mat-BAF) track together with a copy-number values (logR) track are displayed both for SNP array and scGBS data. A legend is provided in the bottom right of the displayed haplarithm profiles in case of disomy and/or deviations from the disomic state from the maternal information according to the principles of haplarithmisis in Figure S1. In both families haplotyping was performed with maternal grandparents and thus, only the maternal haplarithm can be displayed. For the first embryo biopsy (E09 of family 1), interpretation of the logR profile identified a monosomy for chromosome 14 and a segmental trisomy for chromosome 16. In the second embryo biopsy, embryo 8 of family 2, a single trisomy for chromosome 19 is present. Copy-number analysis with a combination of haplarithm and logR profiles allows to specify parental origin for the aberrations. In case of disomic chromosomes, red and blue lines for maternal and/or paternal haplarithms are spaced with a distance of 0.5 apart. Here, in the first embryo, the presence of a maternal monosomy for chromosome 14 (only the maternal copy is remaining) is indicated by a distance of 0 between the red and blue lines of the maternal haplarithm. The segmental trisomy for chromosome 16 is of paternal origin, which is indicated indirectly by the increased distance (= 0.67) between the maternal red and blue lines, which represents a lower fraction of maternal compared to paternal chromosomes. For the second embryo, a meiotic maternal trisomy can be elucidated, since a decreased distance of 0.33 on the maternal haplarithm can be seen. The red and blue lines are centered around 0.5, which indicates the presence of two different haplotypes along the chromosome and hence, corresponds with a meiosis I error. The aberrations were concordant between scGBS and SNP array.