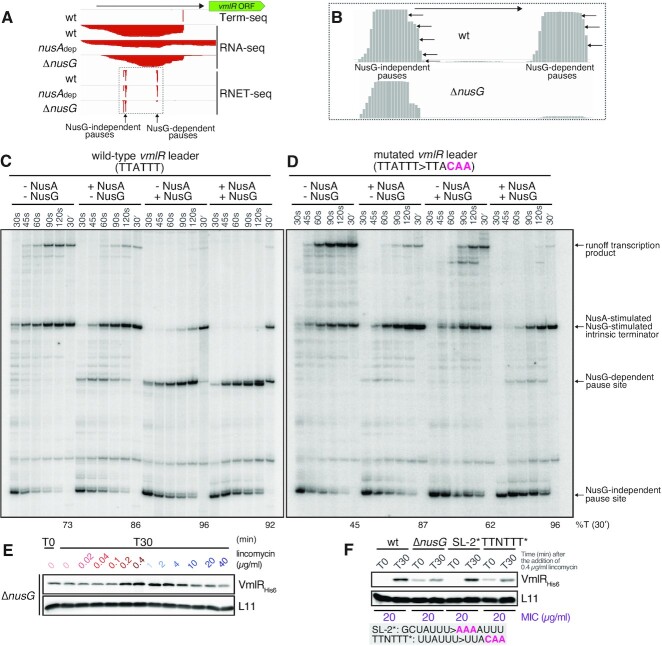
Figure 4.
NusG controls lincomycin-dependent induction of VmlR expression. (A, B) Term-seq and RNA-seq (3,47) as well as RNET-seq (3) in wild-type (PLBS730 grown with IPTG), nusAdep (PLBS730 grown without IPTG), and ΔnusG (PLBS731 grown with IPTG) strains as indicated; ectopic expression of nusA is controlled by an IPTG-inducible Phy-spankpromoter. (A) The intrinsic terminator in the vmlR leader region requires both NusA and NusG for efficient termination. The NusG-dependent and NusG-independent pauses are marked. The arrow at the top indicates the direction of transcription. (B) Zoomed in magnification showing the NusG-dependent and NusG-independent pauses. (C, D) In vitro reconstitution of NusG-dependent RNAP pausing and NusA/NusG-stimulated termination that controls VmlR expression. In vitro transcription termination assays using DNA templates with wild-type (TTATTT, C) or mutant (TTACAA, (D) NusG pause motifs were performed in the absence or presence of NusA, NusG, or both. Mutations in red are in the consensus NusG-dependent pause motif. Percent termination is shown below each of the 30 min chase reaction lanes. (E) VmlR expression is induced by sub-MIC concentrations of lincomycin and the basal expression of VmlR is repressed by NusG. NusG-deficient (ΔnusG) strain encoding His6-tagged VmlR (VHB489) was treated with increasing concentrations of lincomycin for 30 min (T30) and VmlRHis6 was detected by α-His6 immunoblotting. (F) NusG recognizes the TTNTTT motif in the vmlR leader region to repress VmlR expression. Wild-type (VHB223), ΔnusG (VHB489), SL-2* (VHB612) and TTNTTT* (VHB611) strains were treated with 0.4 μg/ml lincomycin for 30 min (T30) and levels of VmlRHis6 were assessed by α-His6 immunoblotting. Mutations of SL-2* and TTNTTT* are highlighted in red.