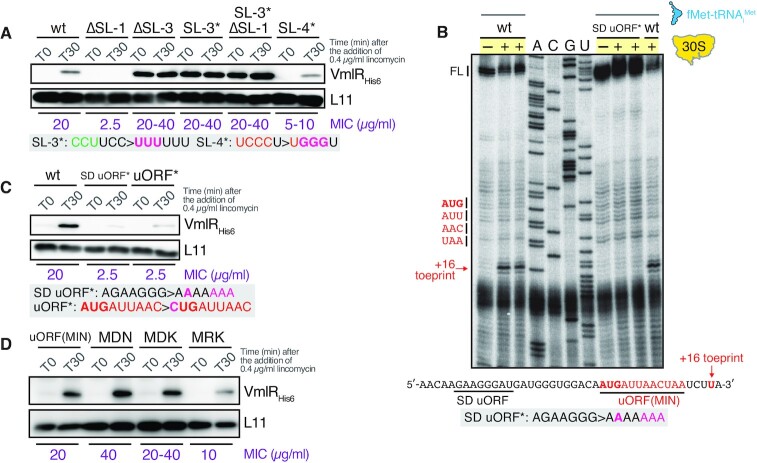
Figure 5.
Lincomycin-dependent induction of VmlR expression is caused by ribosomal stalling at the uORF(MIN). (A) The effects of truncations/mutations in stem loops SL1, SL3 and SL4 on lincomycin-dependent induction of VmlR expression. SL-1 is essential for lincomycin-dependent induction of VmlR expression and SL-3 is crucial for repression of VmlR expression in the absence of the inducer. Wild-type (VHB570), ΔSL-1 (VHB575), ΔSL-3 (VHB474), SL-3* (VHB572), SL-3* ΔSL-1 (VHB583) and SL-4* (VHB594) cells were treated with lincomycin for 30 min and VmlRHis6 expression was detected by immunoblotting. Mutation sites of SL-3* and SL-4* are in red. (B) 30S ribosomal toeprint of the uORF(MIN). Toeprints were conducted using wild-type (wt) and Shine-Dalgarno sequence mutant (SD uORF*) templates as indicated. Reactions were carried out in the absence (–) or presence (+) of B. subtilis 30S ribosomal subunits. All reactions contained E. coli fMet-tRNAifMet. The start codon (AUG), stop codon (UAA), the position of the 30S ribosomal subunit-dependent toeprint located 16 nucleotides downstream the A residue of the start codon, as well as the full-length reverse transcription product (FL) are marked. Sequencing lanes (A, C, G and U) are shown. (C) Translation of the uORF(MIN) is essential for lincomycin-dependent induction of VmlR expression. MIC for lincomycin for wild-type (VHB570), SD uORF* (VHB573) and uORF* (VHB604) strains are indicated in purple. Mutation sites of SD uORF* and uORF* are in red. All experiments were performed at least twice yielding similar results. Expression of VmlRHis6 was assessed by α-His6 immunoblotting. (D) uORF sequence variants from other Bacillus species are competent in induction of vmlR expression in a lincomycin-dependent manner. Wild-type B. subtilis uORF(MIN) (VHB570), MDN (VHB839), MDK (VHB840) and MRK (VHB841) uORF variant cells were treated with lincomycin for 30 min and expression of VmlRHis6 was detected by α-His6 immunoblotting.