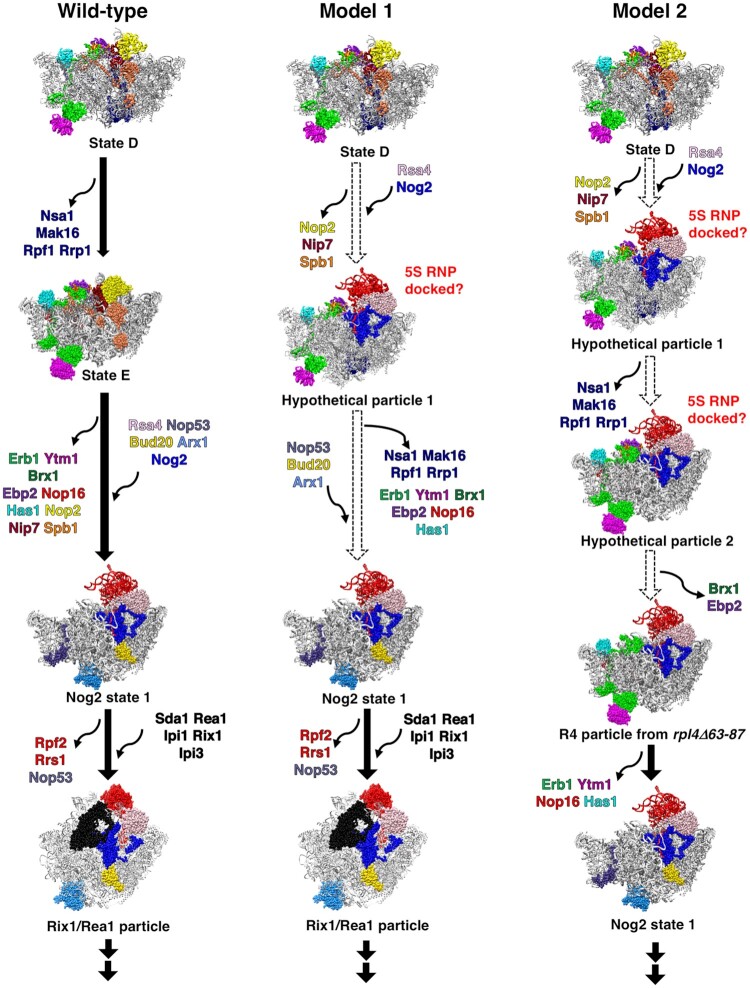
Figure 8.
Hypothetical alternative pathways during assembly of the yeast 60S r-subunit. The left panel represents the standard model for the 60S r-subunit assembly pathway in wild-type cells, while the middle and right panels represent two different models for potential alternative pathways. The thick black arrows represent the sequential order of assembly under wild-type conditions. Dashed white arrows represent hypothetical alternative pathways based on the iTRAQ data from the rpl39Δ mutant. Models for early AFs from atomic models of states D and E were superimposed onto atomic models of Nog2 state 1 or Rix1/Rea1 particles. Particles NE1 and NE2 were omitted for simplification. All hypothetical structures were labeled as such. RNA, and non-relevant r-proteins and AFs are colored grey. For simplicity, helices H68-H69 and H75-H79 were included in the R4 particle, although their densities are missing from the cryo-EM density map of R4 particle. Additionally, potential conformational changes in RNA structures in any of the hypothetical particles were not presented in this model. PDB IDs 6EM5, 6ELZ, 3JCT, and 5FL8 were used for this figure.