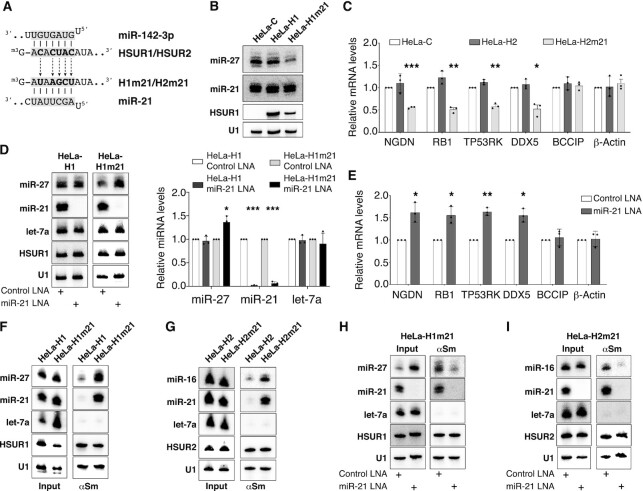
Figure 7.
The identity of the miRNA binding at HSURs’ 5′ end is not important for allosteric regulation. (A) Partial sequences of the 5′ end of HSUR1 and HSUR2, and miR-142-3p and miR-21 seed regions are shown, with residues involved in basepairing highlighted in grey. Mutated residues are shown in bold. (B) Northern blot analysis of equal amount of total RNA prepared from HeLa-C, HeLa-H1, or HeLa cells expressing a mutant version of HSUR1 predicted to bind miR-21 at its 5′ end (HeLa-H1m21). (C) HSUR2 target mRNA levels in HeLa-C, HeLa-H2 and HeLa-H2m21 cells. (D) Same as in (B) on total RNA prepared from HeLa-H1 and HeLa-H1m21 cells transiently transfected with Control LNA inhibitor or an LNA inhibitor with complementarity to miR-21. Right, quantification of independent experiments (n = 3). U1 snRNA signal was used for normalization. (E) Same as in (C) in HeLa-H2m21 cells transiently transfected with Control LNA inhibitor or an LNA inhibitor with complementarity to miR-21. (F) Same as in Figure 3 on extracts prepared from equal number of HeLa-H1 or HeLa-H1m21 cells. (G) Same as in (F) on extracts prepared from equal number of HeLa-H2 or HeLa-H2m21 cells. (H) Same as in Figure 3 on extracts prepared from HeLa-H1m21 cells transiently transfected with Control LNA inhibitor or an LNA inhibitor to miR-21. (I) Same as in (H) on extracts prepared from HeLa-H2m21 cell.