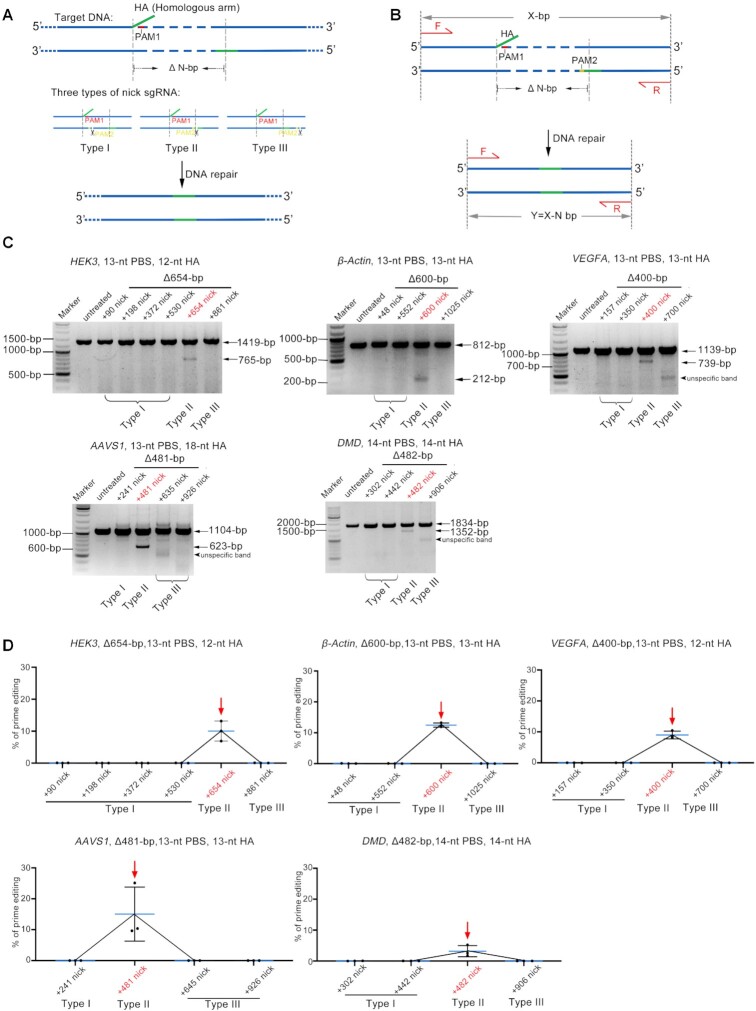
Figure 1.
Positioning nick sgRNA nearby the HA region improve PE3 mediated deletion of large DNA fragment. (A) Schematic diagram showing the design of five types of nicks sgRNAs (Types I, II and III) and the putative editing process of PE3 mediated deletion of large DNA fragment. Homologous arm (HA, homologous to the DNA sequence downstream of the fragment to be deleted) and its complementary region were shown in green. PAMs of pegRNA and nick sgRNA were shown in red and yellow respectively. (B) Schematic diagram showing the detection of targeted deletion by PCR analysis. Paired primers were designed to amplify the targeted deletion and its flanking sequences (fragment without deletion = X; fragment with deletion = Y (Y = X – N). (C) Representative agarose gel electrophoresis detecting the presence of targeted deletion. Note that only type II nicks generated targeted deletions in all three loci studied. (D) Adobe Photoshop CC (2019) quantifying the efficiencies of the targeted deletions. Values and error bars reflect mean ± s.d. of n = 3 independent biological replicates. Data of each replicate were shown in Supplementary Table S5.