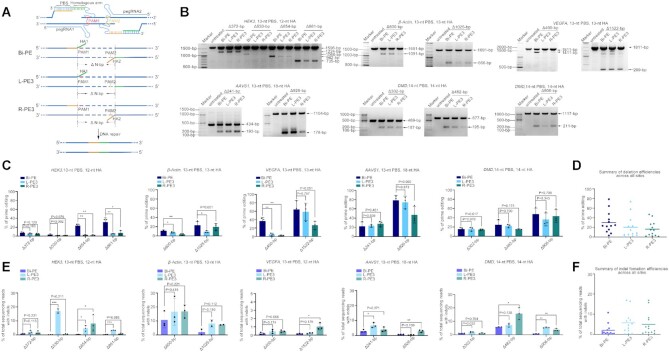
Figure 2.
Bi-PE strategy significantly increased the efficiency of large fragment deletion. (A) Schematic diagram showing the design and the putative editing process of Bi-PE strategy. In Bi-PE strategy, both sgRNAs were designed to be pegRNAs, each of which contained a HA homologous to the sequences proximally flanking the deleted fragment. PE3 containing up-stream pegRNA and down-stream nick sgRNA was designated L-PE3 and vice versa was R-PE3. HA in up-stream pegRNA and its homologous region were shown in green, and the corresponding ones in downstream pegRNA were shown in yellow. (B) Agarose gel analysis of the presence of targeted deletions on indicated loci. (C) Quantification of the efficiency of targeted deletions using Adobe Photoshop CC (2019). Values and error bars reflect mean ± s.d. of n = 3 independent biological replicates. Data of each replicate were shown in Supplementary Table S5. (D) Summary of deletion efficiencies across 5 endogenous sites in HEK293T cells. The mean of all individual values of n = 3 independent biological replicates was plotted. (E) HTS analysis of the frequencies of undesired Indels. The fragments containing deletions were gel purified and amplified by HTS primers. Then the products were subjected to HTS to analyze undesired Indels. All alleles observed with frequency ≥0.01% were shown. Values and error bars reflect mean ± s.d. of n ≥ 2 independent biological replicates. (F) Summary of indel frequencies across five endogenous sites in HEK293T cells. The mean of all individual values of n ≥ 2 independent biological replicates was plotted.