Figure 3.
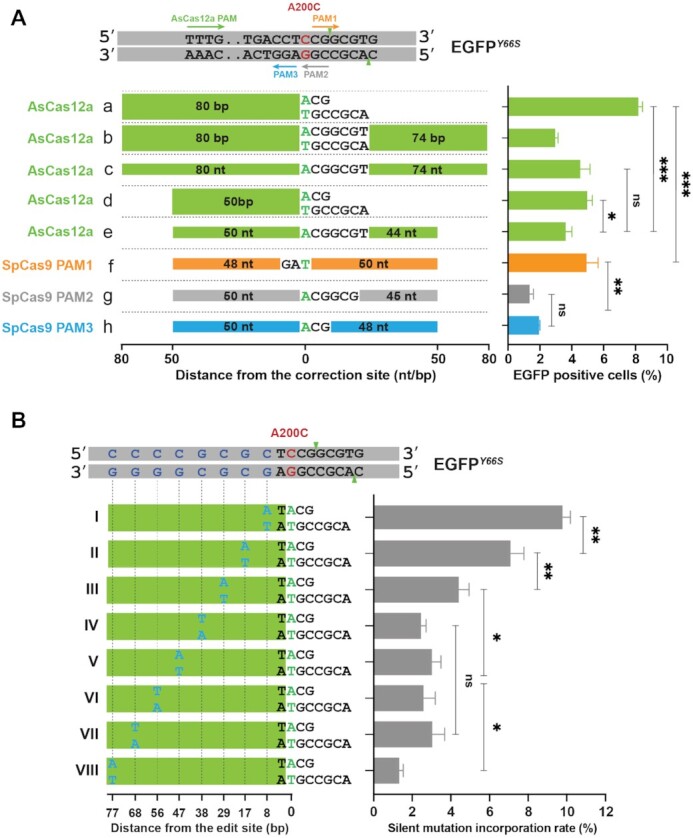
Characterization of LAHR. (A) Comparison between LAHR and HDR in repair of the EGFPY66Sreporter. Cells were targeted using AsCas12a RNP and different repair templates as indicated, (a) a LAHR template with an 80-bp homologous arm and a compatible sticky end; (b) a 160-bp dsDNA template; (c) a 160-nt ssODN template; (d) a LAHR template with a 50-bp homologous arm; (e) a 100-nt ssODN template. For the SpCas9 targeting, the following repair templates were used: (f) a 100-nt ssODN template for PAM1; (g) a 100-nt ssODN template for PAM2; (h) a 100-nt ssODN template for PAM3. Templates (g) and (h) were the same sequence, and the template (f) was their reverse complement sequence. For all repair templates from (a) to (f), the homologous arms are presented as coloured boxes, and different colours indicated different PAM usages. The numbers in the boxes indicate the size of the homologous arm. The correction base (A or T) or base pair (A/T) was in green uppercases. The scale beneath repair templates indicated the distance between each end and the correction site. Indicated percentage of EGFP-positive cells was determined by flow cytometry (See also supplementary figure S12). Error bars correspond to the standard deviation of the average of n = 3 parallel samples. The experiment was repeated three times and a representative dataset is presented here. Statistical test: two-tailed unpaired t-test, ns P > 0.05, * P < 0.05, ** P < 0.01, *** P < 0.001. (B) The repair templates (I–VIII) contained the same A/T base pair (green uppercase) to correct the A200C mutation. A single base pair change (blue uppercases), introducing a silent mutation, on each template was distributed along the homologous arm (green box). The scale at bottom indicates the distance between the silent-mutation-inducing substitution and the repairing A/T base pair. Shown is the percentage incorporation of each silent mutation determined by NGS analysis. Error bars indicate the standard deviation of the average of n = 3 parallel samples. Statistical test: two-tailed unpaired t-test, ns P > 0.05, * P < 0.05.
