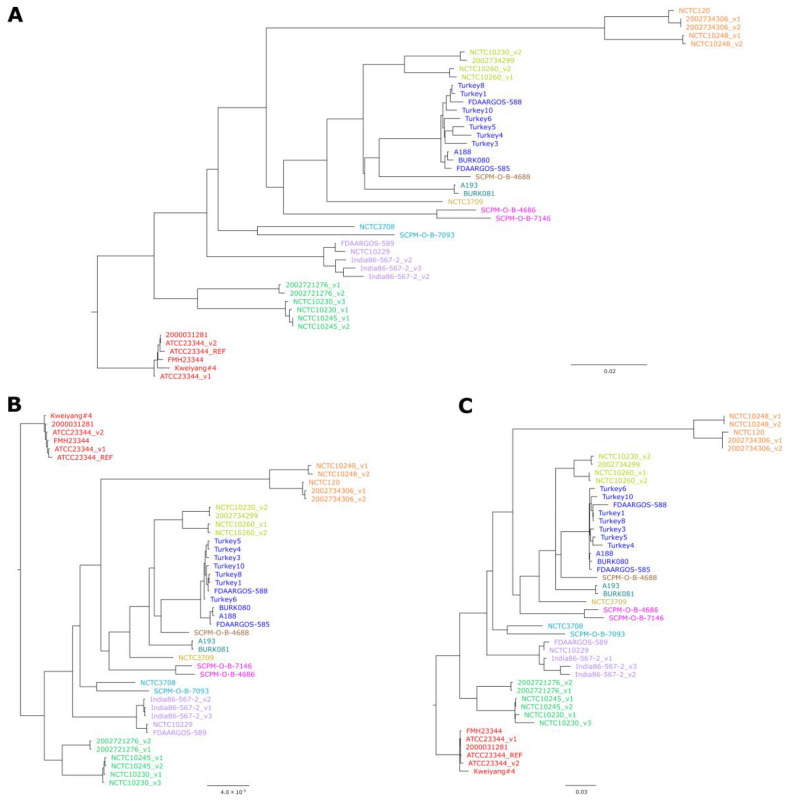
Figure 2.
Comparison between trees generated by different approaches: (A) Neighbor-Joining tree based on cgMLST allelic profiles using 2838 target genes; (B) Maximum-likelihood tree based on cgSNP alignment generated by Snippy using Illumina read data; (C) Approximately Maximum-likelihood tree based on cgSNP alignment generated with Parsnp using genome assemblies. For convenience, clusters or singletons of strains that showed in the trees were colored identically. Bars indicate allelic changes (A) or base substitutions per site (B,C). Bootstrap values for (B,C) can be found in Figure S3.