Fig. 1. A CRISPR gene editing strategy produces stable hPSCs completely lacking cilia.
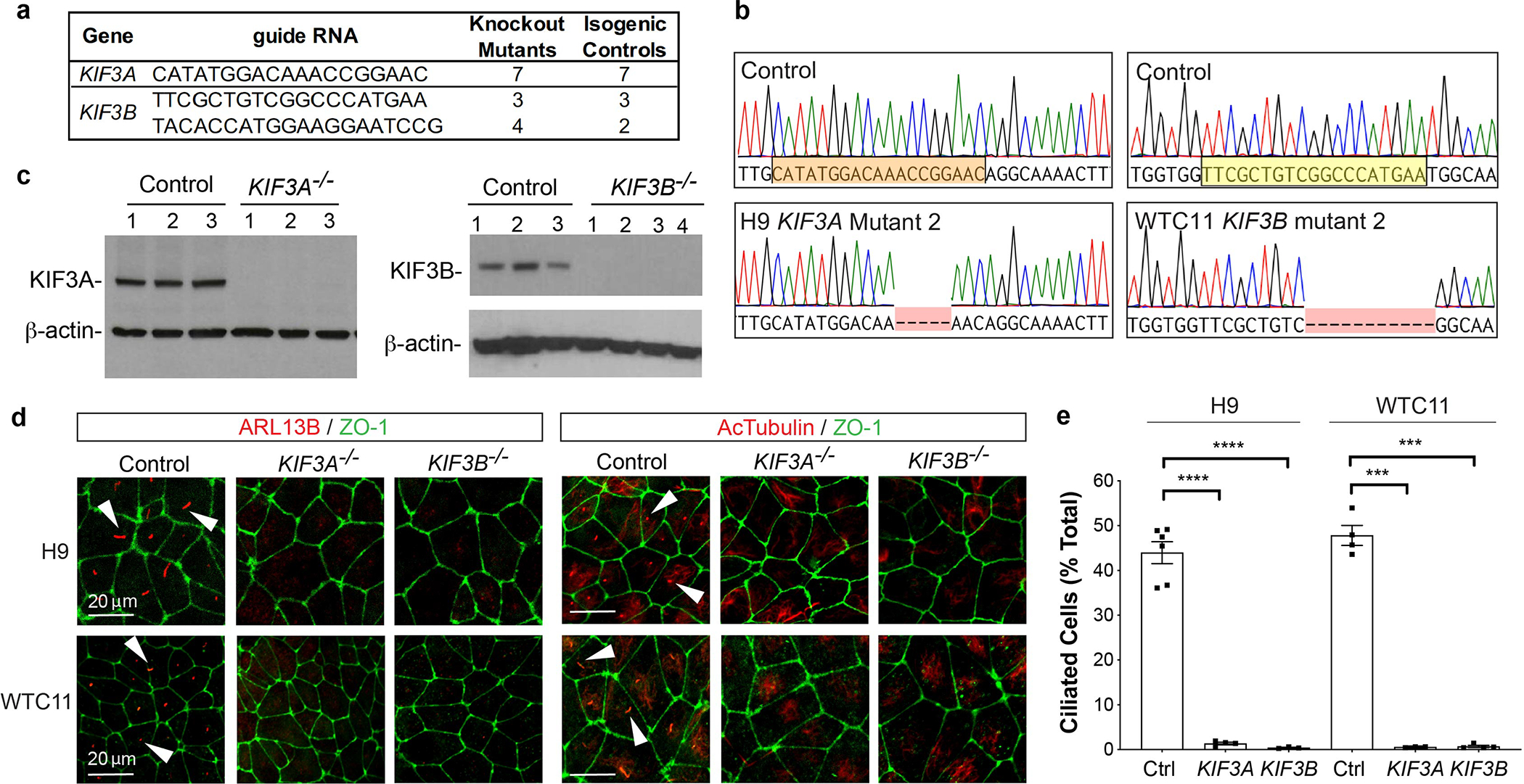
(a) Summary of new hPSC lines. (b) Representative KIF3A and KIF3B chromatograms. Two genetic backgrounds (H9 and WTC11) were used to generate isogenic pairs. The sgRNA sequence used for CRISPR targeting is highlighted. (c) KIF3A and KIF3B immunoblots showing multiple distinct control, KIF3A−/−, and KIF3B−/− hPSC lines. (d) Confocal immunofluorescence images of ARL13B and acetylated α-tubulin (AcTub) with ZO-1 counterstain in representative fields of undifferentiated control and kinesin-2 knockout hPSCs, on two distinct genetic backgrounds (H9 ES cells or WTC11 iPS cells). Representative ciliary axonemes are indicated with arrowheads. (e) Cilia counts in undifferentiated control and kinesin-2 hPSCs on two distinct genetic backgrounds (mean ± s.e.m., n ≥ 3 independent biological replicates per condition from a total of 24 distinct cell lines; ≥ 180 cells counted per cell line). Scale bars, 20 μm. ****, p < 0.0001; ***, p < 0.001.
