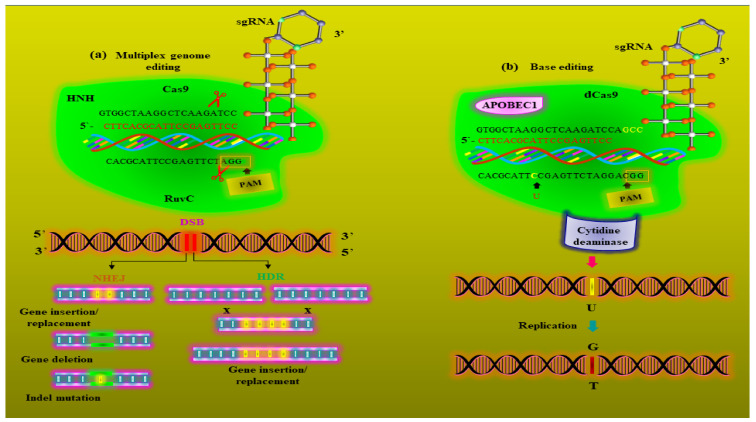
Figure 3.
Schematic representation of how metabolic engineering/editing is done in plants through multiplexed genome editing and multiplexed base editing systems. (a) The CRISPR/Cas9-mediated multiplexed genome editing system consists of multiple single-guide RNAs (gRNAs), and the Cas9 protein is activated via trans-activating CRISPR RNA (tracrRNA) and guided by CRISPR RNA (crRNA) to generate site-specific double-standard breaks (DSBs) at different points on the DNA. The gRNAs detect a unique sequence of 20 nucleotides (red) and the Cas9/gRNAs complex cuts the DNA at a protospacer adjacent motif (PAM) site that is three bases upstream of the target sequence via the RuvC and HNH domains. The DSBs can be repaired either through a homology-directed repair pathway (HDR) or nonhomologous end-joining (NHEJ). (b) shows the modern base-editing system which can be used to edit multiple bases in different pathways for precise metabolic editing. It comprises dead Cas9 (dCas9), which is connected with cytidine deaminase (light blue). The dCas9 is guided by gRNA to target desire single base (yellow) in the DNA sequence and substitute it with another base (brown) distal to the PAM.