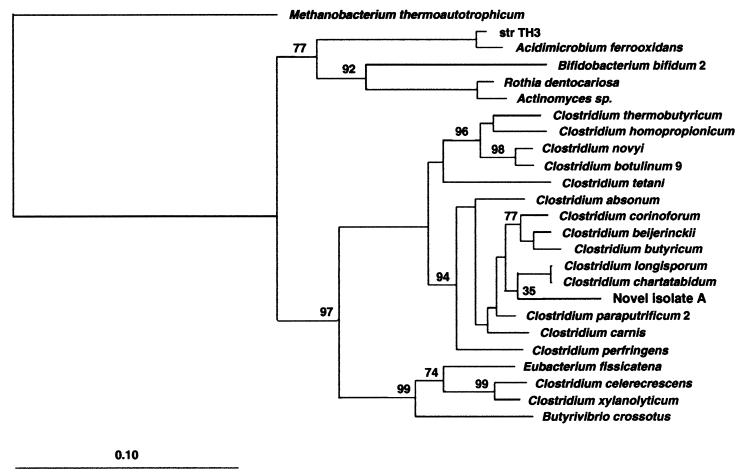
FIG. 5.
Phylogenetic tree of the novel starch-degrading clostridium with other representatives of class I clostridia based on SSU rRNA sequences. The tree was constructed using the neighbor-joining method and the correction of Jukes and Cantor (19). The scale bar indicates 0.10 estimated substitutions per nucleotide. The DNABOOT program of PHYLIP (phylogeny inference package) was used to calculate the bootstrap values, placing confidence limits on phylogenies by using parsimony. Methanobacterium thermoautotrophicum was used to root the tree. Bootstrap values are based on the percentages of 100 replications.