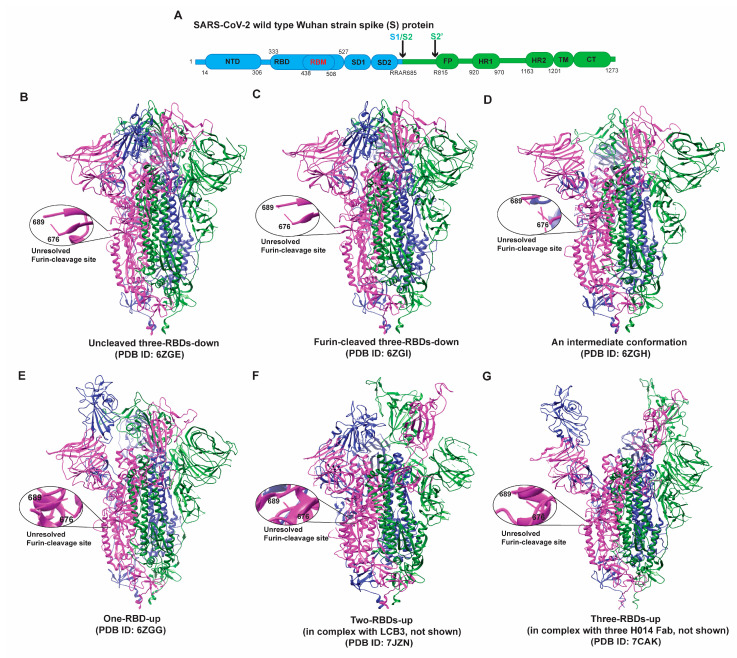
Figure 1.
(A) Schematic map of the full-length (1273 amino acids) Wuhan-Hu-1 or USA/WA1 SARS-CoV-2 Spike protein. S1 and S2 subunits are represented in different colors. S1 subunit comprises the N-terminal domain (NTD), receptor-binding domain (RBD), receptor–binding motif (RBM) within RBD, subunit domain 1 (SD1), and subunit domain 2 (SD2), followed by S1/S2 cleavage site. S2 subunit contains S2′, a protease cleavage site, followed by fusion peptide (FP), heptad repeat 1 (HR1), heptad repeat 2 (HR2), transmembrane domain (TM), and cytoplasmic tail (CT). (B–G) Representation of S structures in different conformational states, including but not limited to an uncleaved, closed ((B), PDB ID 6ZGE), a furin-cleaved, closed ((C), PDB ID 6ZGI), an intermediate ((D), PDB ID 6ZGE), the one-RBD-up state ((E), PDB ID 6ZGG), the two-RBDs-up (in complex with LCB3) ((F), PDB id: 7JZN), and the three-RBDs-up (in complex with three H014 Fabs) ((G), PDB ID 7CAK). The furin cleavage site (682-RRAR-685) at S1/S2 was disordered and thus not resolved in the structures (B–G), as shown in the zoomed-in windows. There is no detectable structural difference (Figure S1) between un-cleaved closed (B) and furin-cleaved closed (C) conformations.