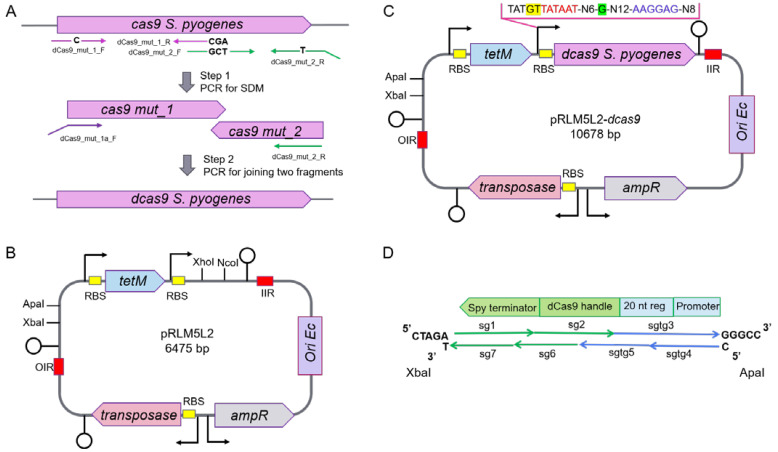
Figure 1.
Schematic of the construction of components for CRISPRi (A) The dcas9 gene was derived from wild-type cas9 S. pyogenes though site-directed mutagenesis (SDM) in two steps: firstly, we changed Asp-10 and His-840 codons to alanyne codons (GAT → GCT and CAC → GCT, respectively), and also TGA was replaced to TAA stop codon using PCR; in the second step, two fragments were linked to full-length dcas9. Primers are indicated by arrows; letters indicate substitute nucleotides. (B) pRLM5L2 transposon vector scheme. Promoters are shown as arrows, terminators–as circles, RBSs are yellow, OIR and IIR-inverted repeats for the integration transposon into the genome. (C) Vector map of the resulting pRM5L2-dcas9. Structure of promoter and RBS for dcas9 expression in mycoplasmas are shown. The EXT element is highlighted in yellow, −10 box are red, initiator nucleotide is green, RBS is blue. (D) Strategy for assembly of dsDNA fragments coding sgRNAs. Each fragment consisted of seven overlapping oligonucleotides, four of which were common to all sgRNAs-coding fragments and three were unique for each specific sgRNA. All oligonucleotides were phosphorylated, ligated together and the resulting product was cloned between the ApaI and XbaI site in pRLM5L2-dcas9. Common oligonucleotides are green arrows, unique are blue. Sticky ends for cloning dsDNA fragments between ApaI and XbaI pre-cut pRLM5L2-dcas9 are highlighted with letters. 20 nt reg is a base-pairing region of coding sgRNA. All oligonucleotides used in this study are listed in Supplementary Table S1.