Table 1.
DNA Binders 1–8 Used for Method Development
| Name | Structure | Binding mechanism | Solubility in aqueous solution | Distinct UV absorbance | Theoretical m/z in MS |
|---|---|---|---|---|---|
|
| |||||
| 9-aminoacridine (1) |
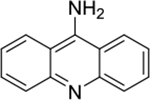
|
intercalator | poor | yes | 195.09 |
| ellipticine (2) |
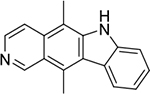
|
intercalator | poor | yes | 247.12 |
| methapyrilene (3) |
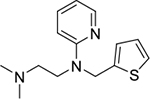
|
intercalator | poor | yes | 262.14 |
| chlorpheniramine (4) |
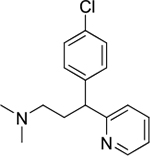
|
intercalator | good | yes | 275.13 |
| bisenzimide (H 33258) (5) |
|
groove binder | poor | yes | 425.21 |
| neomycin (6) |
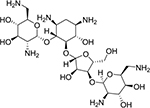
|
groove binder | good | N/A | 615.32 |
| melphalan (7) |
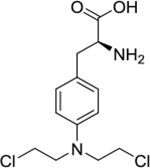
|
covalent binder | poor | yes | 305.08 |
| carmustine (8) |
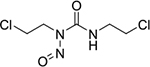
|
covalent binder | poor | yes | N/A |
