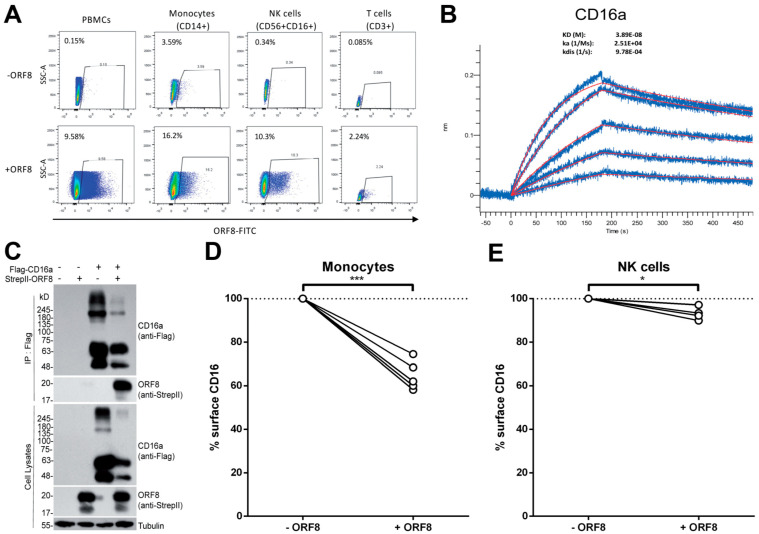
Figure 1.
ORF8 binds monocytes and NK cells through CD16a. (A) PMBCs were incubated with or without FITC-conjugated recombinant ORF8 protein on ice for 30 min, followed by staining with anti-CD14-V450, anti-CD16-PE-CY7 and anti-CD56-PE antibodies before being fixed with 4% PFA and analyzed by flow cytometry. CD3+ T cells were labeled with an anti-CD3-PE antibody. (B) AR2G biosensors loaded with CD16a protein were soaked in two-fold dilution series of ORF8 (31.25 nM–500 nM). Raw data are shown in blue and model in red. The dissociation/affinity constant (KD), on rates (Ka) and off rates (Kdis) were calculated using a 1:1 binding model. (C) HEK293T cells were transfected with CD16a and ORF8 plasmid DNA, followed by co-immunoprecipitation with an anti-Flag antibody. The presence of ORF8 was detected with the anti-StrepII antibody. (D,E) PBMCs from different donors were thawed and incubated for 16 h with media from ORF8 DNA-transfected HEK293T cells. Alternatively, PBMCs were treated with an identical volume of conditioned media collected from HEK293T cells transfected with a control plasmid. The following day, the PBMCs were stained with anti-CD3, anti-CD14, anti-CD19, anti-CD56, anti-CD16 and LIVE/DEAD Fixable Aqua Dead Cell Stain and analyzed by flow cytometry to measure surface levels of CD16 on (D) monocytes (n = 5) and (E) NK cells (n = 4). Cell-surface CD16 levels in presence of ORF8 were normalized on cell-surface CD16 detected in absence of ORF8. Statistical significance was evaluated using a parametric paired t-test. *, p < 0.05; ***, p < 0.001.