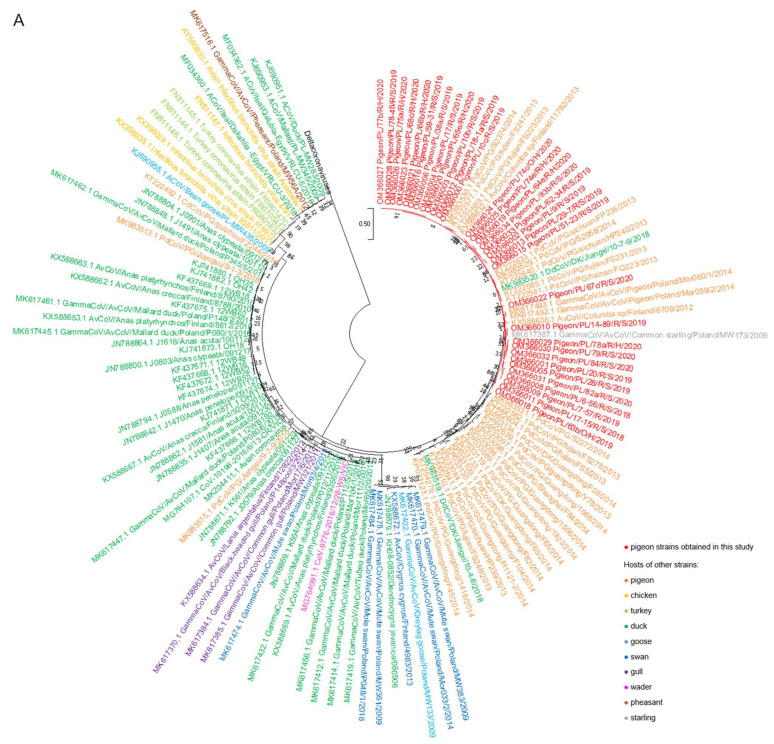
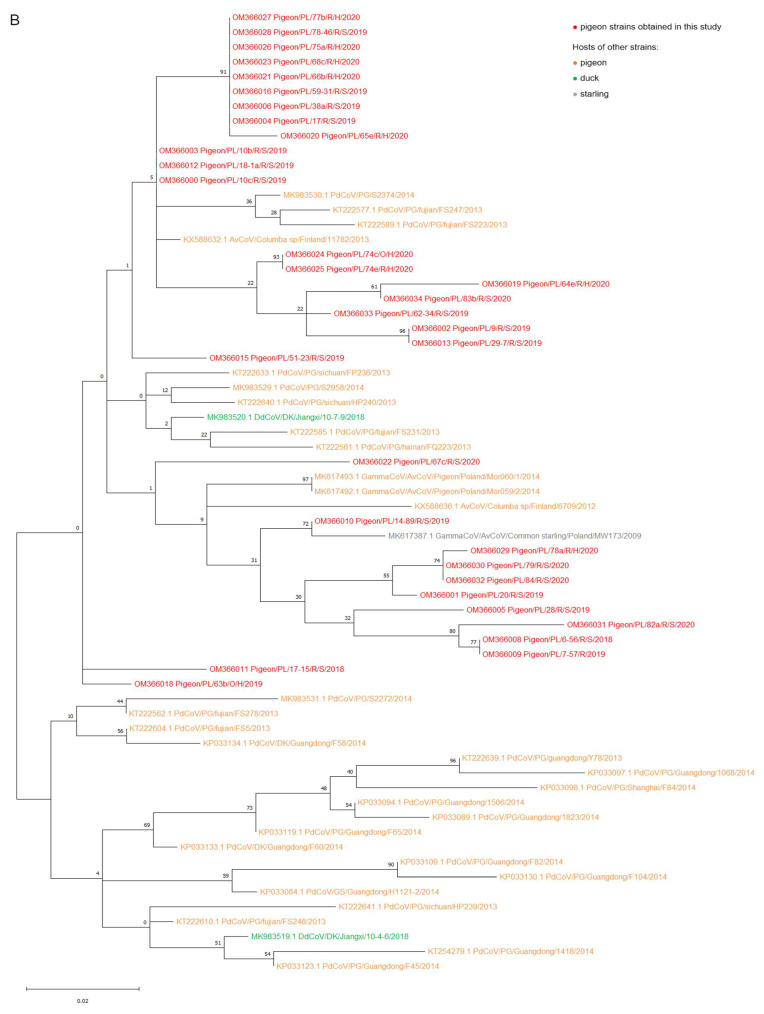
Figure 1.
(A) The phylogenetic tree of RdRp gene nucleotide sequences (440 bp) of 101 various avian coronavirus strains available in the GenBank database and 32 pigeon coronavirus strains obtained in this study. (B) The phylogenetic subtree consisting of the “pigeon-dominant” strains. The trees were inferred in Molecular Evolutionary Genetics Analysis software, version 11 (MEGA 11; https://megasoftware.net/, accessed on 17 December 2021), by using the maximum likelihood method and Tamura 3-parameter model with the highest log likelihood (−4391.32) [28,29]. The initial tree for the heuristic search was obtained automatically by applying the neighbor-joining algorithm to a matrix of pairwise distances estimated using the Tamura 3 parameter model. A discrete gamma distribution was used to model evolutionary rate differences among sites (five categories (+G, parameter = 0.5384)). The bootstrap method was used in a number of 1000 repetitions. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site. All positions containing gaps and missing data were eliminated. The coronavirus strains obtained in this study and from the NCBI database are marked with different colors.