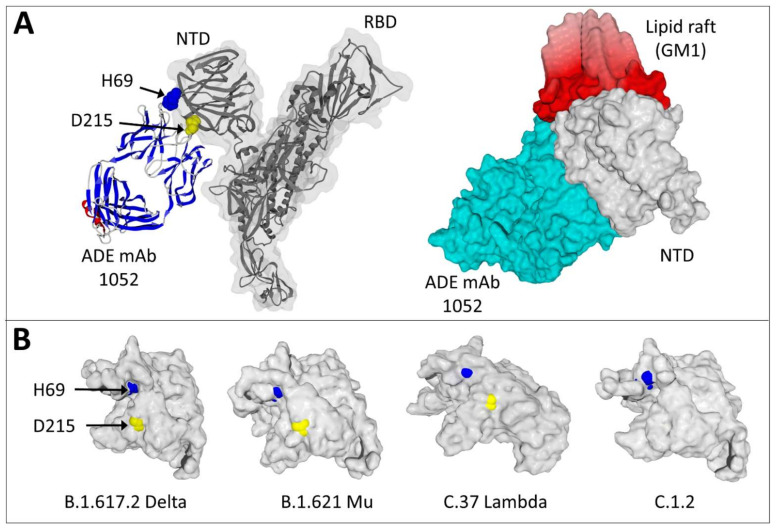
Figure 4.
Critical amino acid residues control the binding of the ADE antibody 1052 to variant spike proteins. (A) Binding of the 1052 antibody (ADE mAb) to the Wuhan spike protein (PDB: 7LAB) with the NTD and RBD indicated. In the left panel, the light and heavy chains of the antibody are represented in standard secondary structures (red, α-helix, blue, β-strand). H69 (in blue atomic spheres) and D215 (in yellow atomic spheres) are highlighted. In the right panel, a surface representation illustrates the geometric complementarity of the Delta spike protein-antibody complex bound to a cluster of gangliosides GM1 figuring a lipid raft on the plasma membrane of a host cell. Note that the 1052 antibody b simultaneously binds to the NTD of the spike protein and to the edge of the lipid raft. (B) Molecular modeling of the NTD of several SARS-CoV-2 variants showing different levels of surface exposure of H69 (in blue) and D215 (in yellow) amino acid residues.