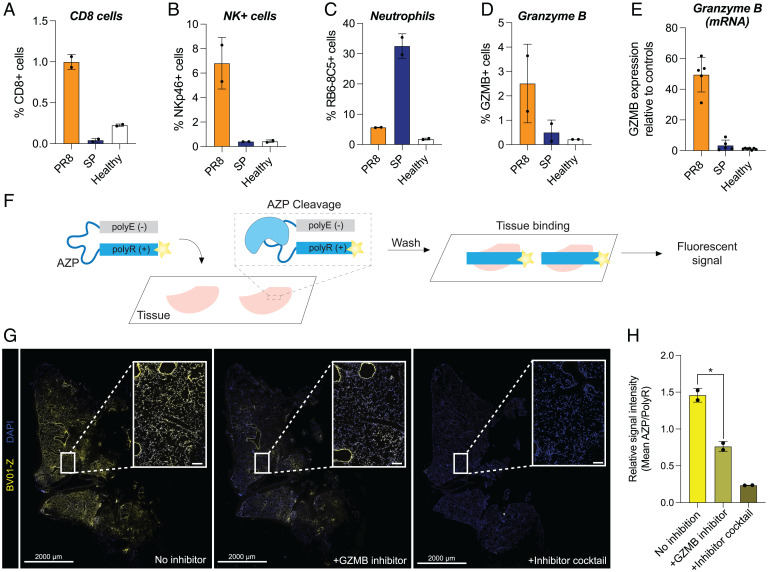
Fig. 5.
GZMB is elevated in viral pneumonia and contributes to nanosensor signal. (A–D) Percentage of detected cells positive for various cell markers via immunofluorescent staining performed on fresh-frozen sections from mice infected with either PR8 or SP (n = 2 consecutive sections per group, mean ± SD). Counts were obtained using QuPath, and stain-positive cells were identified via manually set thresholds. Counts for positive/total cells per section in each panel are (A) CD8 (PR8: 884/83153, 747/80149; SP: 49/86308, 34/127753; healthy: 252/117485, 289/122812); (B) NKp46 (PR8: 4256/80149, 6895/83153; SP: 345/86308, 505/127753; healthy: 593/117485, 421/122812); (C) RB6-8C5 (PR8: 5021/90127, 5549/97327; SP: 38217/128902, 51305/145035; healthy: 2641/130332, 2032/128009); and (D) GZMB (PR8: 1127/82380, 3432/94145; SP: 168/119890, 767/89157; healthy: 335/156523, 291/135411). (E) Relative expression of GZMB in lung tissue from healthy control mice and those with PR8 or SP via qRT-PCR. (F) The original BV01 substrate was incorporated into the AZP BV01-Z, consisting of the substrate sequence linking a fluorescently labeled polyR domain and a polyE domain. The AZP is applied to fresh-frozen tissue and is cleaved by active tissue-resident enzymes, after which the liberated polyR domain electrostatically binds to the tissue. (G) The GZMB responsive AZP BV01-Z (yellow) was applied to PR8-infected tissue with and without a GZMB-specific inhibitor or a broad-spectrum mixture of protease inhibitors. Sections were costained with a free polyR binding control (not shown) and counterstained with DAPI (blue). Staining shows one section of the slide, with white squares marking the location of the inset images. Scale bars for full sections: 2,000 μm; scale bars for zoomed regions: 100 μm. (H) Quantification of relative BV01-Z intensity in sections stained with or without inhibitors (n = 2 consecutive sections, mean ± SD; one-way ANOVA with multiple comparisons and Brown-Forsythe and Welch’s correction, *P = 0.0278).