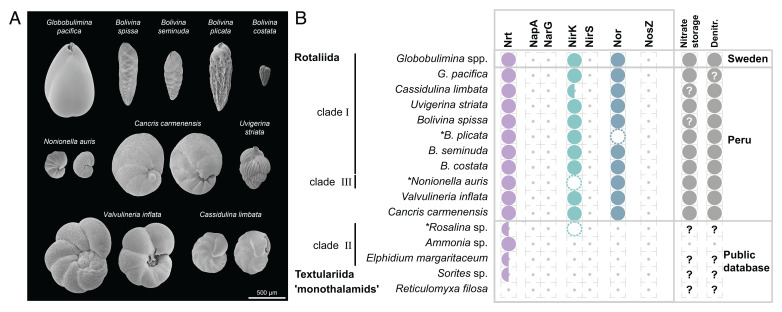
Fig. 1.
Morphological characteristics of the sampled rotaliids and denitrification gene repertoire. (A) Scanning electron micrographs of the sampled species. Species with clearly distinct lateral views are shown from two sides. For more detailed views see SI Appendix, Fig. S1. (B) Presence/absence of homologs of denitrification proteins identified for different foraminifera. Half circles indicate species where only a single, of multiple subtypes, was found. Dashed circles illustrate homologs discarded due to low coverage, and asterisks highlight corresponding species. Evidence for NO3− storage or denitrification activity is illustrated by closed circles in the last column. Question marks denote missing information. Foraminifera sampled in this study are highlighted by the location “Peru.” Protein symbols: Nrt, nitrate/nitrite transporter; NapA, periplasmic nitrate reductase; NarG, membrane-bound nitrate reductase; NirK, copper-containing nitrite reductase; NirS, cd1-containing nitrite reductase; Nor, nitric oxide reductase; NosZ, nitrous oxide reductase. Note that the identification of homologous genes is based not only on sequence similarity but also on transcript abundance in order to exclude bystander species in the data. The additional data filtration stage affected our findings for Rosalina sp., B. plicata, and N. auris (Datasets S2 and S3), where the presence of at least one the crucial homologs (i.e., NirK or Nor) remained in the B. plicata and N. auris metatranscriptomes.