Significance
Steroid hormones control sexual maturation and reproduction in insects and humans alike. The insect steroid hormone ecdysone uses a membrane transporter named Ecdysone Importer (EcI) to enter cells and promote these physiological processes, but EcI is unexpectedly missing in mosquito genomes. Using the yellow fever mosquito Aedes aegypti, here we show that mosquitoes use alternative ecdysone importers to facilitate ecdysone-dependent development and reproduction. These transporters are also present in other insects, including fruit flies, but they are dispensable for fly development and reproduction likely due to their limited expression patterns. Our results thus indicate that differential expression of steroid hormone importers enables tissue- and stage-specific hormone responses, and some importers can obtain critical physiological functions only in certain species.
Keywords: ecdysone, organic anion-transporting polypeptide (OATP), Aedes aegypti, Drosophila melanogaster, vitellogenesis
Abstract
The primary insect steroid hormone ecdysone requires a membrane transporter to enter its target cells. Although an organic anion-transporting polypeptide (OATP) named Ecdysone Importer (EcI) serves this role in the fruit fly Drosophila melanogaster and most likely in other arthropod species, this highly conserved transporter is apparently missing in mosquitoes. Here we report three additional OATPs that facilitate cellular incorporation of ecdysone in Drosophila and the yellow fever mosquito Aedes aegypti. These additional ecdysone importers (EcI-2, -3, and -4) are dispensable for development and reproduction in Drosophila, consistent with the predominant role of EcI. In contrast, in Aedes, EcI-2 is indispensable for ecdysone-mediated development, whereas EcI-4 is critical for vitellogenesis induced by ecdysone in adult females. Altogether, our results indicate unique and essential functions of these additional ecdysone importers in mosquito development and reproduction, making them attractive molecular targets for species- and stage-specific control of ecdysone signaling in mosquitoes.
Ecdysone and other ecdysteroids are a group of steroid hormones that control various aspects of insect development and reproduction (1). Once released into the hemolymph, ecdysone (more specifically, its active form 20-hydroxyecdysone or 20E and related ecdysteroids) enters its target cells to bind to a nuclear receptor named the ecdysone receptor (EcR), which forms a heterodimer with another nuclear receptor Ultraspiracle and induces gene expression (2–5). Although it has long been assumed that ecdysone can pass through the cell membrane through simple diffusion, we recently demonstrated that an organic anion-transporting polypeptide (OATP), which we named Ecdysone Importer (EcI), is required for cellular uptake of ecdysone in the fruit fly Drosophila melanogaster (6). EcI orthologs are found among a wide variety of insects (6), and it is likely that its critical function as a mediator of ecdysone signaling is highly conserved in other insect species (7).
Mosquitoes are the deadliest disease vectors for humans. The yellow fever mosquito Aedes aegypti is the primary vector for arboviruses, including Zika, yellow fever, chikungunya, and dengue viruses, which are of global health concern due to their rapid increases in the geographical distribution (8, 9). Critical functions of ecdysone signaling in Aedes development and reproduction are extensively investigated (10–13), making it an important molecular target for control agents against this deadly virus vector. Surprisingly, however, even in well-annotated mosquito genomes (14–19), an EcI ortholog cannot be found, suggesting the existence of an additional membrane transporter(s) for ecdysone.
In this study, we identified additional OATPs that facilitate cellular uptake of ecdysone in Drosophila and Aedes. These additional ecdysone importer-encoding genes, EcI-2, -3, and -4, are dispensable for normal development and reproduction in Drosophila, confirming the predominant role of EcI in flies. In contrast, CRISPR/Cas9-mediated mutagenesis and RNA interference (RNAi)-mediated knockdown experiments in Aedes suggest that EcI-2 is critical for ecdysone-mediated developmental progression, while EcI-4 is most important for vitellogenesis induced by ecdysone in adult females. Collectively, our results indicate unique functions of these additional ecdysone importers in mosquitoes, making them attractive targets for species- and stage-specific control of mosquito ecdysone signaling.
Results
Identification of Additional Ecdysone Importers in Drosophila and Aedes.
In order to identify all OATPs in mosquitoes and related dipteran species, we thoroughly searched for orthologs of Drosophila OATPs in flies (Musca domestica), sand flies (Phlebotomus papatasi), and mosquitoes (Ae. aegypti, Anopheles gambiae, and Culex quinquefasciatus). Our comprehensive BLAST search identified eight OATPs in M. domestica, eight OATPs in P. papatasi, six OATPs in Ae. aegypti, six OATPs in An. gambiae, and seven OATPs in C. quinquefasciatus (SI Appendix, Table S1). Phylogenetic analysis of these dipteran OATPs confirmed absence of EcI orthologs in mosquitoes (Fig. 1A).
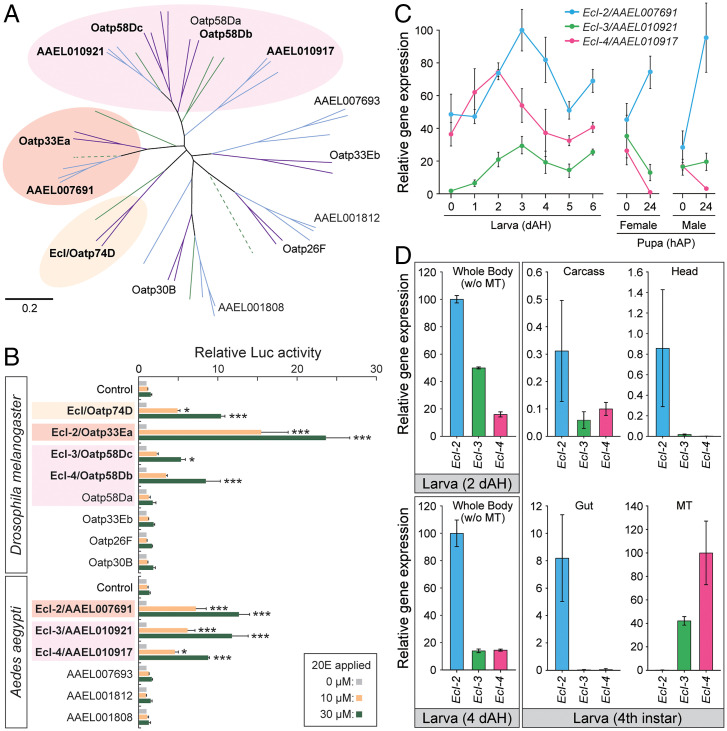
Fig. 1.
Identification of additional ecdysone importers in Drosophila and Aedes. (A) Neighbor-joining unrooted phylogenetic tree of full-length OATP proteins from representative dipteran insect species. Purple, flies (D. melanogaster and M. domestica); green, sand flies (P. papatasi); blue, mosquitoes (Ae. aegypti, A. gambiae, and Culex quinquefasciatus). Dotted lines indicate pseudogenes. Drosophila and Aedes OATPs are labeled. Protein names and GenBank accession numbers are listed in SI Appendix, Table S1. Scale bar indicates an evolutionary distance of 0.2 amino acid substitutions per position. (B) Luciferase (Luc) reporter activities in response to 10 μM or 30 μM 20E in HEK293 cells expressing Drosophila or Aedes OATPs. The cells were transfected with modified EcR (VgEcR) and RXR, along with each OATP-containing vector and luciferase reporter plasmids. Values are relative to the basal level (0 M 20E). All values are the means ± SEM (n = 2 to 4). *P < 0.05, ***P < 0.001 from one-way ANOVA followed by Dunnett’s multiple comparison test as compared to the response of the control cells to the same concentration of 20E. (C) Relative expression levels of ecdysone importer genes in the whole body during Aedes development, as assessed by qRT-PCR. Values are shown as percentages relative to the highest expression level of EcI-2. dAH, days after hatching, hAP, hours after pupation. All values are the means ± SEM (n = 3). (D) Tissue-specific expression of ecdysone importer genes during Aedes development, as assessed by qRT-PCR. Whole body samples without Malpighian tubules from early developmental stages (2 and 4 d after hatching) or individual tissues of fourth instar larvae were collected. Values are shown as percentages relative to the expression levels of EcI-2 in early developmental stages, or EcI-4 in the Malpighian tubule in the fourth instar. MT, Malpighian tubule. All values are the means ± SEM (n = 3).
To identify additional ecdysone importers, we used an ecdysteroid-inducible gene expression system in HEK293 cells (20–22) to test whether the other OATPs in Drosophila and Aedes can facilitate ecdysone signaling in a heterologous system. This led to identification of three additional ecdysone importers in each species: Oatp33Ea, Oatp58Dc, and Oatp58Db in Drosophila, and AAEL007691, AAEL010921, and AAEL010917 in Aedes (Fig. 1B). Based on phylogenetic similarities, these additional ecdysone importers in each species were named EcI-2, -3, and -4, respectively. Each of these OATPs shows ∼60 to 74% amino acid sequence similarities between Drosophila and Aedes.
Temporal expression of EcI-2, -3, and -4 fluctuates during development in mosquitoes and flies (Fig. 1C and SI Appendix, Fig. S1A). With regard to tissue-specific expression, we previously showed that EcI-2 is highly expressed in the gut, whereas EcI-3 and EcI-4 are predominantly expressed in the Malpighian tubules in Drosophila larvae (6). Consistent with this, we observed high expression of EcI-2 in the gut and EcI-3 and EcI-4 in the Malpighian tubules, respectively, in Aedes fourth instar larvae (Fig. 1D). Importantly, in addition to its strong expression in the gut, Aedes EcI-2 is expressed at higher levels in the head and carcass as compared to the other ecdysone importers in fourth instar larvae. Predominant expression of EcI-2 outside of the Malpighian tubules was further confirmed in earlier larval stages (Fig. 1D), suggesting its potential ubiquitous function during larval development.
To confirm their ecdysone importer activities in vivo, Drosophila EcI-2, -3, and -4 were ubiquitously expressed in EcI null flies using the Gal4/UAS system and tested whether they can rescue the first instar arrest phenotype caused by loss of ecdysone signaling (6). As shown in SI Appendix, Fig. S1B, all of these additional ecdysone importers significantly rescued the first instar arrest phenotype of EcI null flies, although the rescued animals could only develop into the second instar as compared to further rescue achieved by expression of UAS-EcI. Importantly, the degree of partial rescue observed by ectopic expression was similar among the three additional importers (SI Appendix, Fig. S1B), suggesting that their functions as ecdysone importers are comparable in vivo.
Unique Requirement of Additional Ecdysone Importers during Mosquito Embryogenesis.
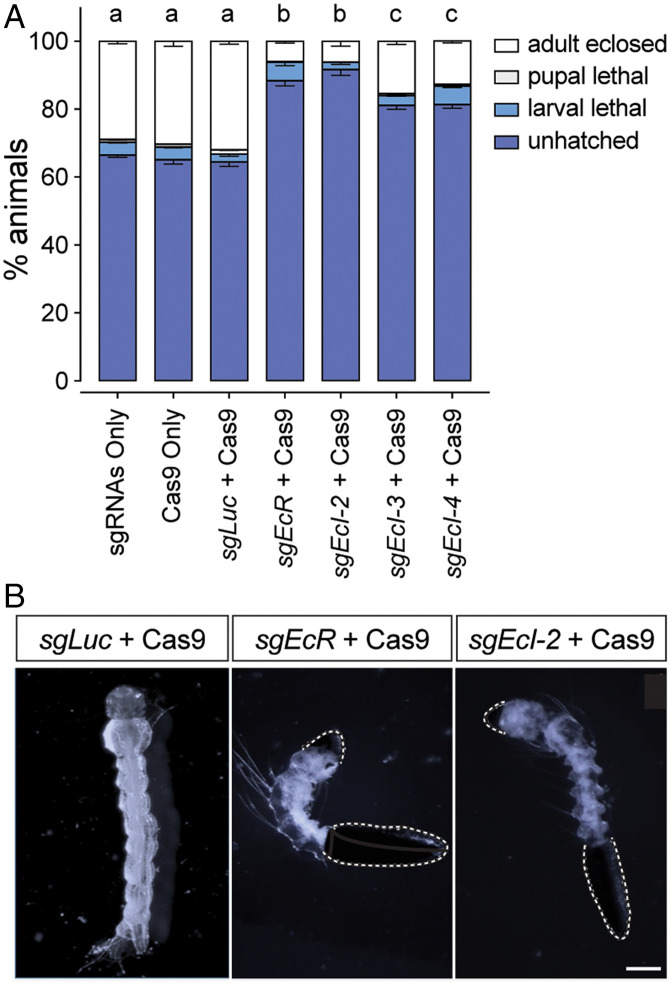
We next conducted CRISPR/Cas9-mediated mutagenesis of EcI-2, -3, and -4 in Drosophila and Aedes, aiming to investigate their functions during the life cycle. In Drosophila, neither individual mutants nor combination mutants showed any discernible developmental or reproductive defects (SI Appendix, Figs. S2 and S3 and Tables S2 and S3), consistent with the role of EcI as the predominant ecdysone importer in flies (6). In contrast, when single guide RNA (sgRNA) for each gene (SI Appendix, Fig. S4) was injected with Cas9 protein into Aedes eggs, all ecdysone importer mutants showed significant embryonic lethality as compared to controls (Fig. 2A). Importantly, only EcI-2 mutants showed levels of embryonic lethality comparable to EcR knockout animals (Fig. 2A). Some of the EcI-2 mutants also showed hatching defects similar to EcR mutants (Fig. 2B), suggesting a critical function of EcI-2 in ecdysone signaling during mosquito embryogenesis.
Fig. 2.
Aedes ecdysone importers are required for embryogenesis. (A) Lethal stages of Aedes embryos injected with Cas9 protein and/or sgRNAs against control (sgLuc), EcR (sgEcR), or ecdysone importer genes (sgEcI-2, sgEcI-3, and sgEcI-4). Total 450 eggs were injected in three independent batches for each treatment, and their lethal stages were monitored thereafter. All values are the means ± SEM. Same letters on Top of the bars indicate statistically insignificant differences in embryonic lethality based on one-way ANOVA with Tukey's honestly significant difference (HSD) test. (B) Hatching defects observed in EcR and EcI-2 mutants. As compared to control (sgLuc + Cas9) that hatched normally, EcR or EcI-2 mutagenesis caused hatching defects in some larvae, where the eggshell remained attached (dotted white lines). (Scale bar, 0.5 mm.)
EcI-2 Is Required for Mosquito Larval Development.
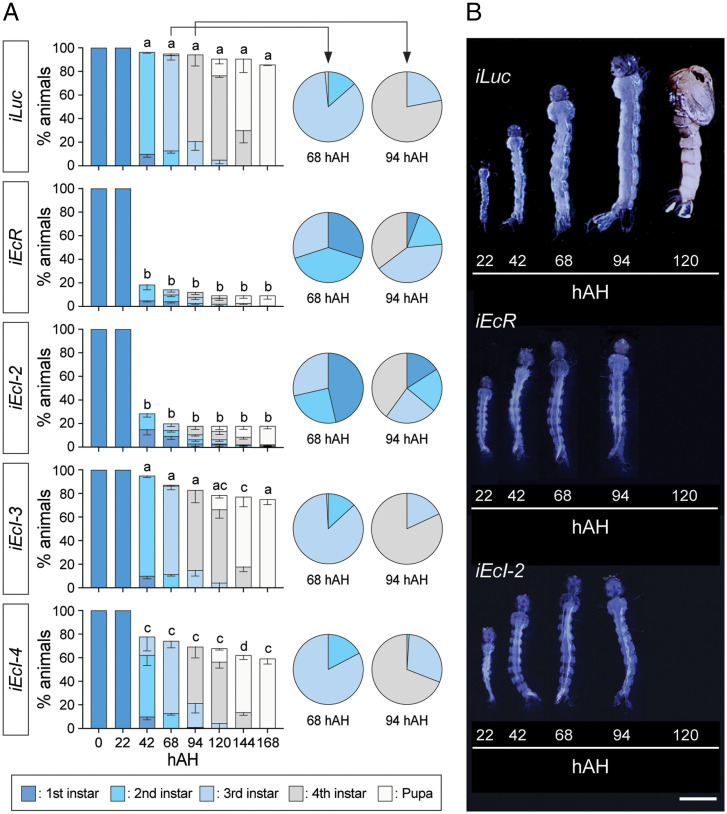
In order to circumvent embryonic lethality in ecdysone importer mutants, we undertook an RNAi approach to further investigate their potential involvement in ecdysone signaling at later developmental stages. Newly hatched Aedes larvae were soaked in double-strand RNA (dsRNA) solutions to suppress expression of each ecdysone importer or EcR, which resulted in ∼50 to 60% reduction of mRNA levels for each target gene (SI Appendix, Fig. S5). Periodic monitoring of their developmental stages and lethality revealed that EcR knockdown induced high (80 to 90%) lethality beginning 42 h after hatching (hAH), consistent with the critical function of ecdysone signaling in molting induction during larval stages (Fig. 3A). Likewise, knockdown of EcI-2, but not EcI-3 or EcI-4, induced high lethality statistically indistinguishable from EcR knockdown during larval development. Importantly, after 68 hAH, many surviving EcR RNAi and EcI-2 RNAi larvae remained as younger instars compared to control animals (Fig. 3A), and their size increase was clearly inhibited (Fig. 3B). This developmental arrest phenotype in EcR RNAi and EcI-2 RNAi animals was further confirmed by detailed measurement of head size and siphon length at two different time points during larval development (SI Appendix, Fig. S6). Furthermore, expression of an ecdysone-inducible gene, E74B, was suppressed by ∼70% in EcI-2 RNAi animals, as compared to milder (∼40%) reduction in EcI-3 or EcI-4 RNAi animals (SI Appendix, Fig. S7). This strong inhibition of E74B expression in EcI-2 RNAi animals was comparable to its suppression induced by RNAi knockdown of EcR (72%) or shade (74%), the gene encoding a cytochrome P450 enzyme required for 20E production (23, 24). Overall, these results further suggest involvement of EcI-2 in ecdysone signaling during mosquito development.
Fig. 3.
Aedes EcI-2 is required for larval developmental transitions. (A) Developmental progression and survival rate (%) of Luc RNAi (iLuc; control), EcR RNAi (iEcR), and EcI-2, -3, and -4 RNAi (iEcI-2, iEcI-3, and iEcI-4) animals. Color bars indicate developmental stages determined by stage-specific morphologic features such as the head capsule size and siphon length (42, 43). All values are the means ± SEM from seven independent experiments with 20 individuals in each replicate. hAH, hours after hatching. Between 42 and 168 hAH, same letters on Top of the bars indicate statistically insignificant differences in survival rate based on one-way ANOVA followed by Bonferroni’s multiple comparison test among RNAi animals at the same time point. Developmental stages of surviving larvae at 68 and 94 hAH are shown on the Right as pie charts. (B) Developmental progression of animals treated with dsRNA targeting Luc (iLuc; control), EcR (iEcR), or EcI-2 (iEcI-2). Representative images of animals at various time points were combined into single panels. (Scale bar, 1 mm.)
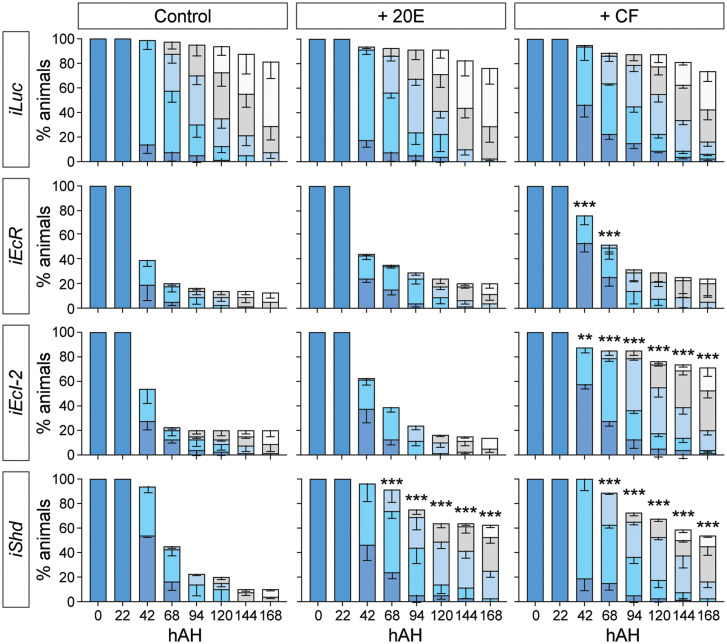
We next attempted to rescue the larval arrest phenotype by administration of two different EcR agonists: the endogenous ecdysteroid 20E and a nonsteroidal insecticide chromafenozide (CF) (Fig. 4). As we have previously shown that CF enters cells independently of EcI (6, 25, 26), we reasoned that CF can activate EcR even in the absence of ecdysone importers, thereby rescuing the developmental defect caused by EcI-2 knockdown. Indeed, CF but not 20E significantly rescued the larval arrest phenotype in EcI-2 RNAi animals, whereas these agonists only partially rescued the larval lethality, if at all, caused by EcR knockdown (Fig. 4). Importantly, both CF and 20E significantly rescued developmental arrest caused by knockdown of shade, confirming the potency of these compounds as EcR agonists in vivo. Collectively, these results indicate that EcR remains functional in the absence of EcI-2, and that EcI-2 is required for 20E to access EcR and induce larval development in mosquitoes.
Fig. 4.
A nonsteroidal ecdysone agonist can rescue developmental arrest caused by EcI-2 knockdown in Aedes. Developmental progression and survival rate (%) of Luc RNAi (iLuc; control), EcR RNAi (iEcR), EcI-2 RNAi (iEcI-2), and shade RNAi (iShd) animals treated with 20E or CF. Color bars indicate developmental stages as shown in Fig. 3A. All values are the means ± SEM from four independent experiments with 20 individuals in each replicate. **P < 0.01, ***P < 0.001 from one-way ANOVA followed by Bonferroni’s multiple comparison test as compared to the survival rate of control (no agonist treatment) of the same RNAi animal at the same time point.
EcI-4 Is Required for Mosquito Vitellogenesis.
In adult female mosquitoes, vitellogenesis after the blood meal is primarily controlled by 20E. 20E up-regulates expression of ecdysone-inducible genes, whose products in turn promote expression of yolk protein precursors such as vitellogenin (Vg) in the fat body (10, 11). When expression levels of ecdysone importers were periodically monitored after the blood meal, we observed significant fluctuation of their expression in all tissues observed, potentially reflecting dynamic changes in ecdysone signaling after blood feeding (SI Appendix, Fig. S8). Importantly, in the abdomen where the fat body is located, expression of EcI-4 was significantly up-regulated as compared to the other ecdysone importer genes between 16 and 20 h after the blood meal (SI Appendix, Fig. S8A). This precedes the peak of ecdysone levels observed ∼18 to 24 h after blood feeding in adult females (27), indicating a critical function of EcI-4 in ecdysone-induced vitellogenesis.
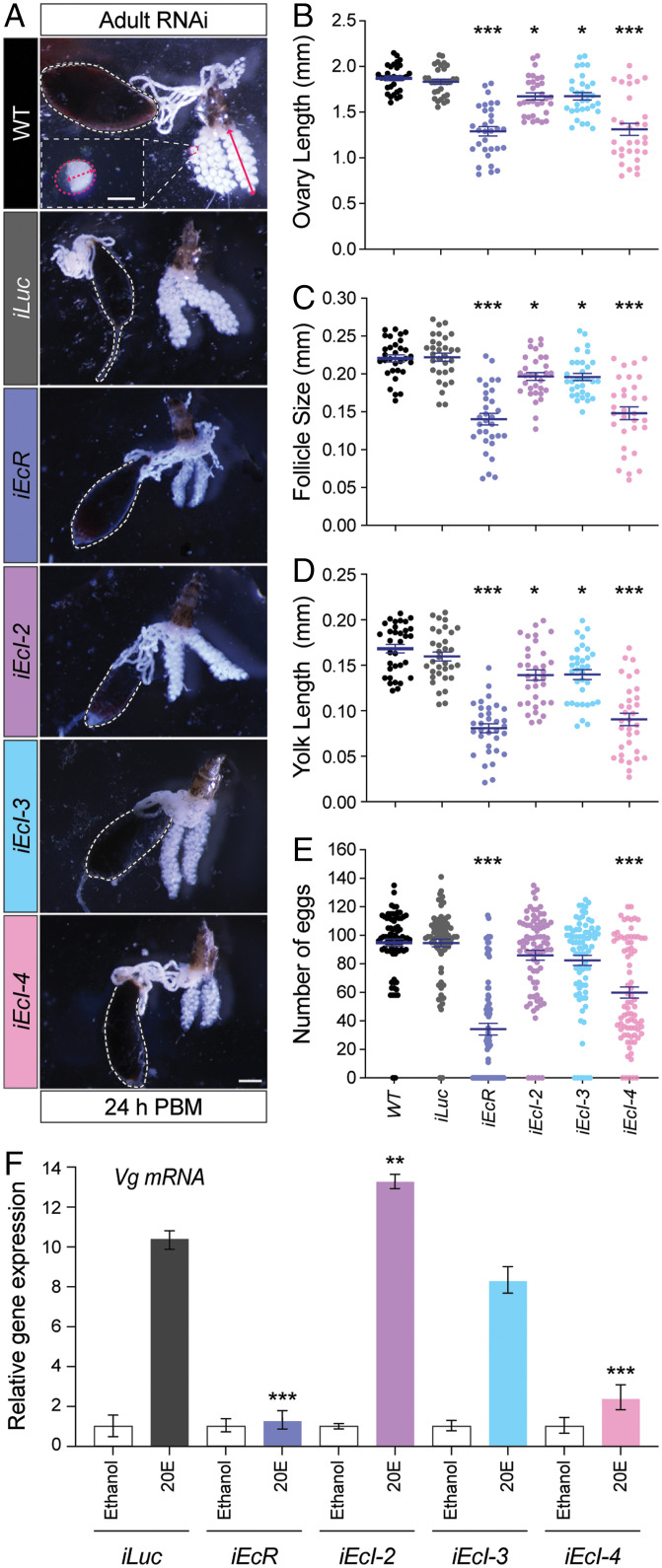
We next injected dsRNA targeting each ecdysone importer or EcR into newly eclosed females, which were blood fed 3 d after injection. This resulted in ∼60 to 75% reduction of mRNA levels for each target gene (SI Appendix, Fig. S9). Vitellogenesis was clearly reduced in EcR RNAi animals as indicated by ovary length, follicle size, and yolk length (Fig. 5 A–D), consistent with the strong reduction of Vg expression in the fat body (SI Appendix, Fig. S9E). Vg expression was also strongly reduced in EcI-4 RNAi animals, as compared to milder reduction caused by EcI-2 or EcI-3 knockdown (SI Appendix, Fig. S9E). Consistent with this, knockdown of EcI-4, but not EcI-2 or EcI-3, caused similar reduction in vitellogenesis comparable to that caused by EcR knockdown (Fig. 5 A–D). As a result, both EcR RNAi and EcI-4 RNAi caused significant reduction in fecundity (Fig. 5E).
Fig. 5.
EcI-4 is required for vitellogenesis in Aedes adult females. (A) Representative images of ovaries from wild-type (WT), Luc RNAi (iLuc; control), EcR RNAi (iEcR), and EcI-2, -3, and -4 RNAi (iEcI-2, iEcI-3, and iEcI-4) females 24 h post blood meal (PBM). The gut filled with the blood meal is surrounded by dashed white lines. At the Top, ovary length, follicle size, and yolk size are indicated by a solid red arrow, dashed red line (longitudinal axis), and dashed red arrow, respectively. (Scale bars, 150 µm for follicle and 0.5 mm for ovary.) (B–D) Ovary length (B), follicle size (C), and yolk length (D) of wild-type (WT), Luc RNAi (iLuc; control), EcR RNAi (iEcR), and EcI-2, -3, and -4 RNAi (iEcI-2, iEcI-3, and iEcI-4) females. All values are the means ± SEM from a minimum of three independent experiments with 10 individuals in each replicate. *P < 0.05, ***P < 0.001 from one-way ANOVA followed by Dunnett’s multiple comparison test as compared to control. (E) Number of deposited eggs per each individual of wild-type (WT), Luc RNAi (iLuc; control), EcR RNAi (iEcR), and EcI-2, -3, and -4 RNAi (iEcI-2, iEcI-3, and iEcI-4) females. All values are the means ± SEM from a minimum of three independent experiments with 10 individuals in each replicate. ***P < 0.001 from one-way ANOVA followed by Dunnett’s multiple comparison test as compared to control. (F) Relative expression levels of Vg in the fat body dissected from Luc RNAi (iLuc; control), EcR RNAi (iEcR), and EcI-2, -3, and -4 RNAi (iEcI-2, iEcI-3, and iEcI-4) females and cultured with or without 10 μM 20E in vitro, as assessed by qRT-PCR. All values are the means ± SEM (n = 4). **P < 0.01, ***P < 0.001 from one-way ANOVA followed by Dunnett’s multiple comparison test as compared to iLuc control.
Lastly, to further confirm that EcI-4 is important in the fat body to mediate ecdysone-induced yolk protein precursor synthesis, we performed in vitro culture experiments using the fat body dissected from RNAi animals. In the control fat body, 20E induced a robust, ∼10-time increase of Vg expression, which was almost completely blocked by EcR RNAi (Fig. 5F). Importantly, knockdown of EcI-4, but not EcI-2 or EcI-3, caused a similar strong inhibition of 20E-induced Vg expression, suggesting a direct role of EcI-4 in ecdysone incorporation into the fat body. Taken together, our results indicate the predominant role of EcI-4 in ecdysone-mediated reproduction in adult female mosquitoes.
Discussion
Recent studies in Drosophila have shown that ecdysone, the primary steroid hormone in insects, requires a membrane transporter EcI to enter its target cells and exert its genomic effects through EcR (6, 25). Although this critical function of EcI in ecdysone signaling seems to be highly conserved among other insect species (7), we noticed that EcI orthologs are unexpectedly missing in mosquitoes. In the present study, we characterized additional ecdysone importers, EcI-2, -3, and -4, in both Drosophila and Aedes. These additional ecdysone importers are dispensable for development and reproduction in Drosophila, likely reflecting the role of EcI as the predominant ecdysone importer. In contrast, in Aedes mosquitoes, EcI-2 is required for ecdysone-mediated developmental progression, whereas EcI-4 seems to be most critical for vitellogenesis in adult females. As these additional ecdysone importers show similar abilities to rescue EcI deficiency in flies, their differential roles in mosquito development and reproduction are most likely due to their distinct expression profiles as revealed in this study. Potential functions of EcI-3 in Aedes mosquitoes are currently unknown.
One of the remaining important questions yet to be fully addressed in ecdysone research is how different types of cells respond differently to a single steroid hormone, even when the cells express similar levels of the receptor. Previous studies have provided a few key answers to this question, including different EcR isoforms (28), cell-specific expression of different EcR cofactors (29, 30), and different chromatin accessibility among different cells (31). Our current study on additional ecdysone importers revealed another important molecular mechanism underlying this issue: These multiple transporters are under control of differential transcriptional regulation, allowing different types of cells to respond to ecdysone at different times. Investigation of such differential regulatory mechanisms of ecdysone importer expression is clearly warranted in future studies.
The evolutionary reason why EcI was lost in mosquito species is currently unknown. Our results nonetheless suggest that, in the absence of EcI, alternative ecdysone importers assume paramount importance in development and reproduction. This makes these additional ecdysone importers potential targets for development of mosquito-specific insect growth regulators and pesticides. The involvement of distinct ecdysone importers in development and reproduction may even make it possible to develop stage-specific regulators of ecdysone signaling in mosquitoes. The effort to identify efficient blockers of ecdysone importers is currently underway.
What are the functions of the additional ecdysone importers, particularly in insects that have EcI in the genome? Although EcI-2, -3, and -4 are dispensable for Drosophila development and reproduction, it is conceivable that they have either unique or redundant functions that are not essential for overall viability and fertility under normal conditions. In this regard, it would be important to examine potential functions of these additional ecdysone importers in Drosophila and other insects more closely under different conditions, particularly in tissues where these transporters are highly expressed (6). Considering pleiotropic functions of ecdysone in insect physiology (1), it would also be interesting to investigate whether these additional ecdysone importers are involved in ecdysone functions that are not directly related to growth and reproduction, such as stress responses. Some tools developed in the current study, such as EcI-2, -3, and -4 mutant flies, are expected to facilitate such future studies. Importantly, OATPs generally have a broad spectrum of substrates, and it has previously been reported, for example, that EcI-4 in the Malpighian tubules is involved in excretion of a plant-derived toxic substance in flies (32). It is also interesting to note here that a recent report identified a potential role of Oatp33Eb in promoting ecdysone uptake into cachectic tumors in Drosophila (33), although Oatp33Eb did not facilitate ecdysone uptake in our heterologous system. This may indicate functional interactions between multiple OATPs through trafficking of substrates other than ecdysteroids. When investigating as yet unknown functions of ecdysone importers, it is imperative to consider influences caused by transport of such additional substrates.
In conclusion, our results indicate unique functions of newly characterized ecdysone importers in development and reproduction in mosquitoes, which may pave the way for better control of these deadly human disease vectors. Functional characterization of ecdysone importers in different insect species is expected to deepen our understanding of how pleiotropic functions of ecdysone are regulated in different tissues under different conditions.
Materials and Methods
Flies.
All flies (D. melanogaster) were raised at 25 °C on standard fly food (6) under a 12-h light/dark cycle. w1118 was used as a control strain. EcI mutant alleles (EcI1 and EcI2) were generated previously (6); all the other ecdysone importer mutant alleles (EcI-21, EcI-31, EcI-41, and EcI-3–41) were generated using CRISPR/Cas9-mediated mutagenesis as described below. Deficiency alleles over ecdysone importer genes [Df(2L)Exel6033, #7516; Df(2R)Exel7171, #7902] and armadillo (arm)-Gal4 (#1560) were obtained from the Bloomington Drosophila Stock Center (BDSC). UAS-EcI-2, UAS-EcI-3, and UAS-EcI-4 were generated by subcloning full-length cDNA clones into pUAST vector (see below for details of cDNA clones). All new transgenic flies were generated by BestGene Inc.
Mosquitoes.
The Rockefeller strain of Ae. aegypti was used in this study. Both male and female adults were maintained on 10% sucrose and reared at 27 °C and 80% relative humidity under a 16-h light/8-h dark cycle. Larvae in water were fed on a diet containing dry dog food (Blue Buffalo), fish flakes (Tetramin), and liver powder (Now Foods) (10:10:1 wt. ratio). All mosquito dissection was performed in 1× phosphate-buffered saline (PBS) solution (Fisher BioReagents) at room temperature. Using an artificial glass feeder, all adult female mosquitoes were allowed to feed on bovine blood purchased from Hemostat. Only fully engorged female mosquitoes were used.
Comprehensive Identification of OATPs in Dipteran Species.
Using amino acid sequences of all eight Drosophila OATPs as queries, whole-genome sequences of the housefly Musca domestica, the sand fly P. papatasi, the yellow fever mosquito Ae. aegypti, the African malaria mosquito An. gambiae, and the southern house mosquito C. quinquefasciatus were screened (TBLASTN analysis) using NCBI BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and VectorBase BLAST (https://vectorbase.org/vectorbase/app/search/transcript/UnifiedBlast). All the obtained sequences were reciprocally screened against Drosophila proteome (BLASTP analysis) by using FlyBase BLAST (https://flybase.org/blast/) to confirm their orthologous relationships with Drosophila OATPs. This two-step screening identified eight OATPs in M. domestica, eight OATPs in P. papatasi (two of which are likely pseudogenes), six OATPs in Ae. aegypti, six OATPs in An. gambiae, and seven OATPs in C. quinquefasciatus. Protein names and GenBank accession numbers are listed in SI Appendix, Table S1. The sequence similarities between Aedes and Drosophila ecdysone importers were calculated using the EMBOSS Water Pairwise Sequence Alignment tool (https://www.ebi.ac.uk/Tools/psa/emboss_water/) (34).
Phylogenetic Tree Analysis.
The unrooted neighbor-joining tree (Fig. 1A) was generated using ClustalW (https://www.ddbj.nig.ac.jp/services/clustalw-e.html) (35, 36). Entire amino acid sequences of all OATPs in the dipteran species mentioned above (SI Appendix, Table S1) were aligned. Bootstrap analyses of 1,000 replications were conducted to assess the relationships.
Cloning of OATP-Encoding Genes.
For Drosophila OATP-encoding genes, full-length cDNA clones from the Drosophila Genomics Resource Center (EcI-2/Oatp33Ea, LD36578; EcI-3/Oatp58Dc, LP09443; Oatp58Da, IP17768; Oatp33Eb, RE09129; Oatp26F, RE32029; and Oatp30B, RE26532) were subcloned into the pcDNA3.1 vector. The coding sequence of EcI-4/Oatp58Db was PCR amplified from whole-body Drosophila cDNA using Phusion Plus DNA Polymerase (Thermo Fisher Scientific) with the primers listed in SI Appendix, Table S4. The PCR product was cloned into the pcDNA3.1 vector and sequenced.
For Aedes OATP-encoding genes, total RNA was collected from fourth instar larvae using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. cDNA was generated from purified total RNA using PrimeScript RT Master Mix (Takara Bio). PCR amplification of the sequence corresponding to EcI-2/AAEL007691, EcI-3/AAEL010921, EcI-4/AAEL010917, AAEL001812, AAEL001808, and AAEL007693 was performed using Phusion Plus DNA Polymerase with the primers listed in SI Appendix, Table S5. The PCR products were cloned into the pcDNA3.1 vector and sequenced.
Transfection and Luciferase Reporter Assay in HEK293 Cells.
HEK cells (obtained from Michael E. Adams, University of California, Riverside, CA) at a density of 4 × 105 cells/mL were seeded in 100 µL/well of Opti-MEM reduced serum media (Thermo Fisher Scientific) containing 5% fetal bovine serum (FBS) and 1% MEM nonessential amino acids (NEAA) solution (Thermo Fisher Scientific) in a 96-well clear flat bottom microplate (Corning). Transfection of HEK293 cells was performed using Attractene transfection reagent (Qiagen) by the fast-forward transfection approach following the manufacturer’s instructions. A total of 50 µL/well of transfection mixture containing Opti-MEM reduced serum media, Attractene, and DNA plasmids was added to each well, bringing the final volume to 150 µL/well. A total of 0.1 µg/well of pcDNA3.1 empty vector (control) or pcDNA3.1 vector containing full-length cDNA of Drosophila or Aedes OATP-encoding genes was transfected, along with 60 ng/well of pERV3 receptor plasmid (Agilent Technologies) containing a modified ecdysone receptor (VgEcR) and RXR, 36 ng/well of pEGSH-LUC luciferase reporter plasmid (Agilent Technologies), and 0.9 ng/well of pRL-CMV Renilla luciferase reporter plasmid (Promega) as a reference. After 24 h of incubation at 37 °C and 5% CO2, transfection medium was removed and replaced with 150 µL/well of Dulbecco's Modified Eagle Medium (DMEM) with 4.5 mg/mL glucose and sodium pyruvate without L-glutamine and phenol red (w-G-SP, wo-G-PR) (Thermo Fisher Scientific) containing 10% FBS, 1% Penicillin-Streptomycin Solution (PSS), and 1% MEM NEAA solution. After 48 h of incubation at 37 °C and 5% CO2, medium was removed and replaced with 150 µL/well of DMEM (w-G-SP, wo-G-PR) containing 1% PSS, 1% MEM NEAA solution, and 20E at indicated concentrations. After 24 h of incubation at 37 °C and 5% CO2, 75 µL of media was removed (total 75 µL remaining per well) and 75 µL/well of Dual-Glo Solution was added (total 150 µL/well). After 20 min of incubation at room temperature in the dark, 120 µL/well of cell lysates were transferred to 96-well solid white flat bottom polystyrene TC-treated plates. The firefly luciferase activity and cotransfected Renilla luciferase activity were measured subsequently using the Dual-Luciferase Reporter Assay System in accordance with the manufacturer’s instructions and analyzed with GloMax-Multi + Microplate Multimode Reader with Instinct. For each condition, cells were treated independently in three wells of a 96-well plate in each experiment, and the same experiment was conducted multiple times on different days.
Total RNA Extraction and qRT-PCR.
Animals or dissected tissues were collected in 1.5-mL tubes and immediately flash-frozen in liquid nitrogen. Total RNA from animals or tissues was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Extracted RNA was further purified by RNeasy mini kit (Qiagen) following the manufacturer’s instructions, combined with treatment with RNase-Free DNase Set (Qiagen). cDNA was generated from purified total RNA using PrimeScript RT Master Mix (Takara Bio). qRT-PCR was performed on the CFX connect real-time PCR detection system (Bio-Rad) using SYBR Premix Ex Taq II (Tli RNaseH Plus) (Takara Bio). For absolute quantification of mRNAs, serial dilutions of pGEM-T (Promega) plasmids containing coding sequences of the target genes or internal control genes (rp49 for D. melanogaster and AeRpL32 (AAEL003396) for Ae. aegypti) were used as standards. After the molar amounts were calculated, transcript levels of the target mRNA were normalized to internal control gene levels in the same samples. Three to four separate samples were collected for each experiment and duplicate measurements were conducted. The primers used are listed in SI Appendix, Tables S4 and S5.
EcI Mutant Rescue Experiments in Drosophila.
EcI mutant rescue experiments were conducted by weak ubiquitous expression of UAS-EcI-2, -3, and -4 transgenes, respectively, in the EcI mutant background. arm-Gal4; EcI1/TM6b-Dfd-EGFP flies were crossed to either w1118 (heterozygous mutant control), EcI2/TM6b-Dfd-EGFP (transheterozygous mutant control), UAS-EcI; EcI2/TM6b-Dfd-EGFP (positive control), UAS-EcI-2; EcI2/TM6b-Dfd-EGFP, UAS-EcI-3; EcI2/TM6b-Dfd-EGFP, or UAS-EcI-4; EcI2/TM6b-Dfd-EGFP, respectively. Eggs were laid on grape juice plates with yeast paste at 25 °C for 6 h. After 24 h, early first instar larvae just after hatching were collected (homozygous mutant larvae were collected by selecting those without the balancer chromosome with GFP) and transferred into vials with standard food (fewer than 50 animals/vial). Developmental stages were scored at 36 h and 120 h after hatching by checking stage-specific morphology of larval mouth hooks and posterior spiracles (6).
Generation of EcI-2, EcI-3, and EcI-4 Mutants in Drosophila.
EcI-2, EcI-3, and EcI-4 mutant alleles were generated using the CRISPR/Cas9 system. Pairs of gRNA target sequences (20 bp: T1 and T2) were designed near the transcription start and stop sites of each target gene using NIG-FLY Cas9 Target finder (NIG) (SI Appendix, Fig. S2). T1 from EcI-3 and T2 from EcI-4 were used for making the EcI-3–4 double mutant. Forward and reverse 24-bp oligonucleotides with 20-bp target sequences (Table S4) were annealed to generate a double-strand DNA with 4-bp overhangs on both ends and inserted into BbsI-digested pBFv-U6.2 or pBFv-U6.2B vector provided by the NIG (37). To construct double-gRNA vectors, the first gRNA (T1) was cloned into pBFv-U6.2 (named pBFv-U6.2-T1), whereas the second gRNA (T2) was cloned into pBFv-U6.2B (named pBFv-U6.2B-T2). A fragment containing the U6 promoter, and the first gRNA was cut out from pBFv-U6.2-T1 and ligated into pBFv-U6.2B-T2 (named pBFv-U6.2B-T1-T2). These double-gRNA vectors (pBFv-U6.2B-T1-T2) were independently injected into embryos of yw;; nos-cas9 (III-attP2)/TM6B flies (BestGene Inc).
For each target gene, surviving G0 males were divided into 10 groups and crossed en masse to Sp/CyO-GFP (obtained from Takashi Nishimura) virgin female flies. From the progeny of each of these 10 crosses, five single males were isolated and crossed independently to Sp/CyO-GFP virgin female flies to establish independent isogenized lines. To confirm deletions of the target genes, we extracted genome DNA from these 50 lines using the DNA extraction buffer (10 mM Tris⋅HCl, 1 mM Ethylenediaminetetraacetic acid, 25 mM NaCl, and 200 µg/mL Proteinase K), and PCR amplification was performed by using primers listed in SI Appendix, Table S4. Out of 50 lines, several lines from each target pair possessed deletion mutations. The PCR products were sequenced, and complete deletions were confirmed in all lines. We selected one mutant allele for each target gene and named them as EcI-21, EcI-31, EcI-41, and EcI-3–41. EcI-21 has a 3,704-bp deletion including the 5′ untranslated region and almost the entire EcI-2/Oatp33Ea coding sequence (CDS) (SI Appendix, Fig. S2A). EcI-31 has a 3,535-bp deletion including the 5′ untranslated region and almost the entire EcI-3/Oatp58Dc CDS (SI Appendix, Fig. S2B). EcI-41 has two deletions with a 9-bp deletion including the transcriptional start site and a 2,092-bp deletion including almost the entire EcI-4/Oatp58Db CDS (SI Appendix, Fig. S2C). EcI-3–41 has a 6,495-bp deletion including the 5′ untranslated region of EcI-3/Oatp58Dc, the entire EcI-3/Oatp58Dc CDS, and almost the entire EcI-4/Oatp58Db CDS (SI Appendix, Fig. S2D).
Pupariation Timing Analysis in Drosophila.
Flies were allowed to lay eggs on 3% agar plates containing 30% grape juice (Welch’s) at 25 °C. Newly hatched larvae were transferred into vials with mashed standard food (25 larvae/vial), and pupae were counted at indicated time points. For each genotype, four vials were scored two times a day.
Pupal Volume Measurements in Drosophila.
Pupal length and width were determined from images captured with a Zeiss Axiocam 506 color digital camera attached to a SteREO Discovery.V12 microscope (Zeiss). Images were processed using ImageJ 1.53v (NIH). The pupal volume was determined by the following approximate equation: 4/3π (length × width2).
Egg Laying Assay in Drosophila.
Four-day-old single virgin females and five to six males were transferred to a vial with standard food to allow mating and egg laying. After 24 h, individual females were then transferred to a fresh vial for further egg laying for another 24 h. The total number of eggs laid in 2 d (48 h) was counted.
Mutagenesis in Aedes.
sgRNAs were designed immediately downstream of the predicted start codon of each ecdysone importer gene (SI Appendix, Fig. S4). One guanine was added at the 5′ terminal end of each sgRNA to facilitate transcription by T7 RNA polymerase (SI Appendix, Table S5). Potential off-target binding was checked by using two online tools: zifit.partners.org/ZiFiT/andcrispr.mit.edu. dsDNA templates for sgRNA synthesis were generated by template-free PCR, using a specific forward primer for each gene and one universal reverse primer (SI Appendix, Table S5). sgRNA was synthesized using the MEGAscript T7 Transcription Kit (Ambion) and purified using the MEGAclear Transcription Clean-Up Kit (Ambion) following the manufacturer’s protocols. A mixture of sgRNA (40 ng/μL) and Cas9 protein with a nuclear localization signal (PAN Bio; 300 ng/μL) was microinjected into the posterior pole of preblastoderm embryos at an angle of 10 to 25°. The sgRNA/Cas9 ratio was optimized according to a previously described protocol (38). The embryos were hatched 5 d postinjection and reared thereafter as described above. The microinjection was repeated three times for a total of 450 embryos per each gene.
The genomic DNA was extracted from newly hatched larvae using the DNA extraction buffer. Fragments containing sgRNA target sites were amplified using gene-specific primers (SI Appendix, Table S5). The PCR products were gel purified using the Gel DNA Recovery Kit (Zymoclean), cloned into the pGEMT-easy vector (Thermo Fisher Scientific), and sequenced. CRISPR-Cas9 mutagenesis efficiency was assessed through a T7 Endonuclease 1 (T7E1) assay. In brief, 200 ng of PCR products in 1× NEB buffer 2 (New England Biolabs) were hybridized under the following conditions: 95 °C for 5 min, 95 to 85 °C at −2 °C/s, 85 to 25 °C at −1 °C/s, and held at 4 °C. Ten units of T7E1 enzyme (New England Biolabs) were added to each sample, and the samples were incubated at 37 °C for 15 min. Products were visualized using 2% agarose gel electrophoresis.
dsRNA Synthesis for RNAi in Aedes.
For dsRNA synthesis, a T7 promoter sequence, TAATACGACTCACTATAGGGAGA, was added to the 5′ end of each primer as listed in SI Appendix, Table S5. PCR was performed using the OneTaq-Quick-Load 2× Master Mix (New England BioLabs) with Aedes whole-body cDNA as a template, and the amplified PCR products were cleaned using the Gel DNA Recovery Kit (Zymoclean). dsRNA was synthesized using the MEGAscript T7 Transcription Kit (Ambion).
RNAi in Aedes Larvae.
Prior to hatching, mosquito eggs were allowed to develop for a minimum of 1 wk. To collect newly hatched larvae, eggs were submerged in a container with water and food and left at room temperature overnight. On the following day, the container was moved into the incubator set at 27 °C and 80% relative humidity. Twenty newly hatched larvae were transferred into a 1.5-mL Eppendorf tube containing 100 µL of premixed food and dsRNA (0.5 µg/µL final concentration). Larvae were soaked in the dsRNA solution for 4 h, after which they were transferred into a 1-oz cup containing 4 mL of premixed food. All surviving larvae were periodically monitored and counted thereafter. The head capsule length (head size) and siphon length were measured using images captured with a Zeiss Axiocam 506 color digital camera attached to a SteREO Discovery.V12 microscope (Zeiss).
For 20E and CF rescue experiments, larvae were cultured in 20E (Sigma-Aldrich; final concentration of 1 µM in 0.004% ethanol), CF (Sigma-Aldrich; final concentration of 10 nM in 0.00002% ethanol), or 0.004% ethanol (control) from 22 h after hatching, and surviving larvae were periodically monitored and counted thereafter.
RNAi in Aedes Adult Females.
Purified dsRNA was resuspended with high-performance liquid chromatography-grade water (Thermo Fisher Scientific) at 5 μg/μL. Newly emerged virgin females were separated using a manual aspirator into a new cage and provided with 10% sucrose. Three days after eclosion, virgin females were anesthetized on ice and injected with 2.0 μg dsRNA (400 nL) using a Nanoject II microinjector (Drummond Scientific Company). After injection, females were placed back in a cage with 10% sucrose to recover for 24 h at 27 °C with 80% humidity.
Ovary Length, Follicle Size, and Yolk Length Measurements in Aedes Adult Females.
After postinjection recovery, adult females were allowed to mate for 48 h and then allowed to feed on bovine blood for 1 h. Fully engorged females were selected and isolated in a cage and provided with 10% sucrose. At 24 h post blood meal, dsRNA-injected females were anesthetized for 5 min at −20 °C. Ovaries were then dissected, and the length of the central longitudinal axis of the ovary was measured. The ovary length of each female was calculated as an average length of the two ovaries. For primary follicle size measurement, the length of the longitudinal axis of 15 primarily follicles per ovary was measured, and the average follicle length of each female was calculated. The yolk length of each female was determined by measuring the average yolk length of all oocytes from one ovary. All measurements were conducted using images captured with a Zeiss Axiocam 506 color digital camera attached to a SteREO Discovery.V12 microscope (Zeiss). Images were processed using ImageJ 1.53v (NIH).
Egg Laying Assay in Aedes.
After postinjection recovery, adult females were allowed to mate for 48 h and then allowed to feed on bovine blood for 1 h. Fully engorged females were selected, and individual females were placed in a cup with oviposition paper. The number of eggs was counted at 96 h post blood meal.
In Vitro Aedes Fat Body Culture.
The fat body culture medium was prepared as previously described (39), with the TES buffer prepared as described in ref. 40. In vitro fat body culture experiments were performed as reported previously (39–41). Fat bodies dissected from five adult females 2 d after dsRNA injection were incubated in a single well of 96-well plates for 6 h in the culture medium with either 10 μM 20E or solvent (0.04% ethanol). After incubation, fat bodies were harvested, and total RNA extraction and qRT-PCR were performed as described above.
Supplementary Material
Acknowledgments
We thank A. S. Raikhel for his generous support in establishing our mosquito research project, the Bloomington Drosophila Stock Center (NIH P40 OD018537), T. Nishimura for fly stocks, M. E. Adams for cell lines, the Drosophila Genomics Resource Center (NIH P40 OD010949), the National Institute of Genetics Fly Stock Center, D. J. Mangelsdorf for vectors and cDNA clones, and A. S. Raikhel and M. E. Adams for critical reading of the manuscript. This study was supported by a Postdoctoral Fellowship for Research Abroad from the Japan Society for the Promotion of Science to N.O., the Naito Foundation Subsidy for Dispatch of Young Researchers Abroad to N.O., an NIH Director’s New Innovator Award DP2 GM132929 to N.Y., a research grant from the W. M. Keck Foundation to N.Y., and a Pew Biomedical Scholars Award from the Pew Charitable Trusts to N.Y.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2202932119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Yamanaka N., Ecdysteroid signalling in insects—From biosynthesis to gene expression regulation. Adv. Insect Physiol. 60, 1–36 (2021). [Google Scholar]
- 2.Thomas H. E., Stunnenberg H. G., Stewart A. F., Heterodimerization of the Drosophila ecdysone receptor with retinoid X receptor and ultraspiracle. Nature 362, 471–475 (1993). [DOI] [PubMed] [Google Scholar]
- 3.Yao T.-P., Segraves W. A., Oro A. E., McKeown M., Evans R. M., Drosophila ultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell 71, 63–72 (1992). [DOI] [PubMed] [Google Scholar]
- 4.Yao T.-P., et al. , Functional ecdysone receptor is the product of EcR and Ultraspiracle genes. Nature 366, 476–479 (1993). [DOI] [PubMed] [Google Scholar]
- 5.Riddiford L. M., Cherbas P., Truman J. W., Ecdysone receptors and their biological actions. Vitam. Horm. 60, 1–73 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Okamoto N., et al. , A membrane transporter is required for steroid hormone uptake in Drosophila. Dev. Cell 47, 294–305.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rösner J., Tietmeyer J., Merzendorfer H., Organic anion-transporting polypeptides are involved in the elimination of insecticides from the red flour beetle, Tribolium castaneum. J. Pest Sci. 94, 1427–1437 (2021). [Google Scholar]
- 8.Kraemer M. U., et al. , The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. eLife 4, e08347 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leta S., et al. , Global risk mapping for major diseases transmitted by Aedes aegypti and Aedes albopictus. Int. J. Infect. Dis. 67, 25–35 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy S., Saha T. T., Zou Z., Raikhel A. S., Regulatory pathways controlling insect reproduction. Annu. Rev. Entomol. 63, 489–511 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Hansen I. A., Attardo G. M., Rodriguez S. D., Drake L. L., Four-way regulation of mosquito yolk protein precursor genes by juvenile hormone-, ecdysone-, nutrient-, and insulin-like peptide signaling pathways. Front. Physiol. 5, 103 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L., Zhu J., Sun G., Raikhel A. S., The early gene Broad is involved in the ecdysteroid hierarchy governing vitellogenesis of the mosquito Aedes aegypti. J. Mol. Endocrinol. 33, 743–761 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Raikhel A. S., Vitellogenesis in mosquitoes. Adv. Dis. Vector Res. 9, 1–39 (1992). [Google Scholar]
- 14.Matthews B. J., et al. , Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature 563, 501–507 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domingos A., Pinheiro-Silva R., Couto J., do Rosário V., de la Fuente J., The Anopheles gambiae transcriptome - A turning point for malaria control. Insect Mol. Biol. 26, 140–151 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Silva Martins W. F., et al. , Transcriptomic analysis of insecticide resistance in the lymphatic filariasis vector Culex quinquefasciatus. Sci. Rep. 9, 11406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neafsey D. E., et al. , Mosquito genomics. Highly evolvable malaria vectors: The genomes of 16 Anopheles mosquitoes. Science 347, 1258522 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharakhova M. V., et al. , Update of the Anopheles gambiae PEST genome assembly. Genome Biol. 8, R5 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arensburger P., et al. , Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science 330, 86–88 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christopherson K. S., Mark M. R., Bajaj V., Godowski P. J., Ecdysteroid-dependent regulation of genes in mammalian cells by a Drosophila ecdysone receptor and chimeric transactivators. Proc. Natl. Acad. Sci. U.S.A. 89, 6314–6318 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.No D., Yao T. P., Evans R. M., Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 93, 3346–3351 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saez E., et al. , Identification of ligands and coligands for the ecdysone-regulated gene switch. Proc. Natl. Acad. Sci. U.S.A. 97, 14512–14517 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petryk A., et al. , Shade is the Drosophila P450 enzyme that mediates the hydroxylation of ecdysone to the steroid insect molting hormone 20-hydroxyecdysone. Proc. Natl. Acad. Sci. U.S.A. 100, 13773–13778 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rewitz K. F., Gilbert L. I., Daphnia Halloween genes that encode cytochrome P450s mediating the synthesis of the arthropod molting hormone: Evolutionary implications. BMC Evol. Biol. 8, 60 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamoto N., Yamanaka N., Steroid hormone entry into the brain requires a membrane transporter in Drosophila. Curr. Biol. 30, 359–366.e3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masterson M., Bittar R., Chu H., Yamanaka N., Haga-Yamanaka S., Rapid assessment of insect steroid hormone entry into cultured cells. Front. Physiol. 12, 816058 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagedorn H. H., et al. , The ovary as a source of alpha-ecdysone in an adult mosquito. Proc. Natl. Acad. Sci. U.S.A. 72, 3255–3259 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schubiger M., Tomita S., Sung C., Robinow S., Truman J. W., Isoform specific control of gene activity in vivo by the Drosophila ecdysone receptor. Mech. Dev. 120, 909–918 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Shlyueva D., et al. , Hormone-responsive enhancer-activity maps reveal predictive motifs, indirect repression, and targeting of closed chromatin. Mol. Cell 54, 180–192 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Stoiber M., Celniker S., Cherbas L., Brown B., Cherbas P., Diverse hormone response networks in 41 independent Drosophila cell lines. G3 (Bethesda) 6, 683–694 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uyehara C. M., et al. , Hormone-dependent control of developmental timing through regulation of chromatin accessibility. Genes Dev. 31, 862–875 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torrie L. S., et al. , Resolution of the insect ouabain paradox. Proc. Natl. Acad. Sci. U.S.A. 101, 13689–13693 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santabárbara-Ruiz P., Léopold P., An P., An Oatp transporter-mediated steroid sink promotes tumor-induced cachexia in Drosophila. Dev. Cell 56, 2741–2751.e7 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Madeira F., et al. , The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47 (W1), W636–W641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitou N., Nei M., The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987). [DOI] [PubMed] [Google Scholar]
- 36.Larkin M. A., et al. , Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Kondo S., Ueda R., Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195, 715–721 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kistler K. E., Vosshall L. B., Matthews B. J., Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. Cell Rep. 11, 51–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chung H. N., et al. , Fat body organ culture system in Aedes aegypti, a vector of Zika virus. J. Vis. Exp. 2017, 55508 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roy S. G., Hansen I. A., Raikhel A. S., Effect of insulin and 20-hydroxyecdysone in the fat body of the yellow fever mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 37, 1317–1326 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roy S., et al. , Regulation of gene expression patterns in mosquito reproduction. PLoS Genet. 11, e1005450 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christophers S. R.,Aedes aegypti (L.), The Yellow Fever Mosquito. Its Life History, Bionomics, and Structure (Cambridge University Press, Cambridge, 1960). [Google Scholar]
- 43.Bar A., Andrew J., Morphology and morphometry of Aedes aegypti larvae. Annu. Res. Rev. Biol. 3, 1–12 (2013). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.