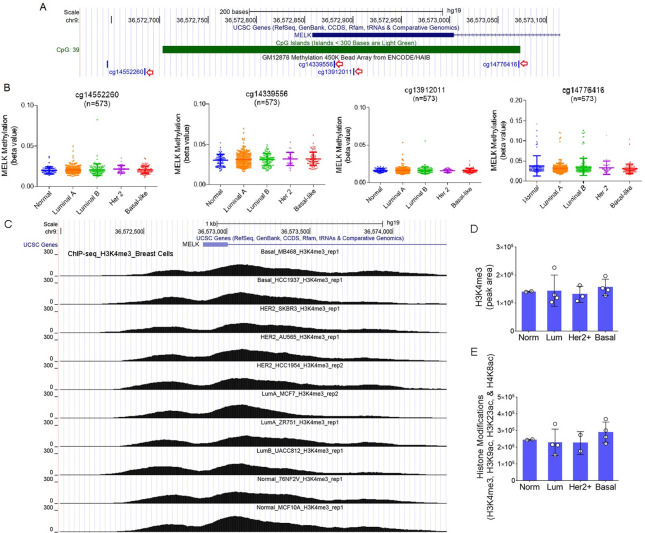
Fig 2. Epigenetic regulation of the MELK promoter in breast tumors.
(A) The UCSC genome browser shows the location of CpG islands in the MELK promoter at chromosome 9p13.2. The four CpG dinucleotides located closest to the transcription start site were selected for analysis and are indicated by red arrows. (B) Analysis of TCGA HumanMethylation450 Array data showed no difference in CpG site methylation among breast cancer subtypes. Beta values (0 to 1) are relative values increasing from hypomethylation to hypermethylation. (C) ChIP-seq data for H3K4me3 in breast cancer cells were visualized through the UCSC genome browser (chr9:36,571,990–36,574,891). Each peak represents the level of H3K4me3 modification in each cell line. (D and E). The levels of H3K4me3 (C) and other histone modifications (H3K4me3, H3K9ac, H3K23ac and H4K8ac combined) (D) were calculated as a total peak area (Y-axis, details in the methods) and compared by molecular subtype. No significant differences in histone modifications were observed among breast cancer subtypes.