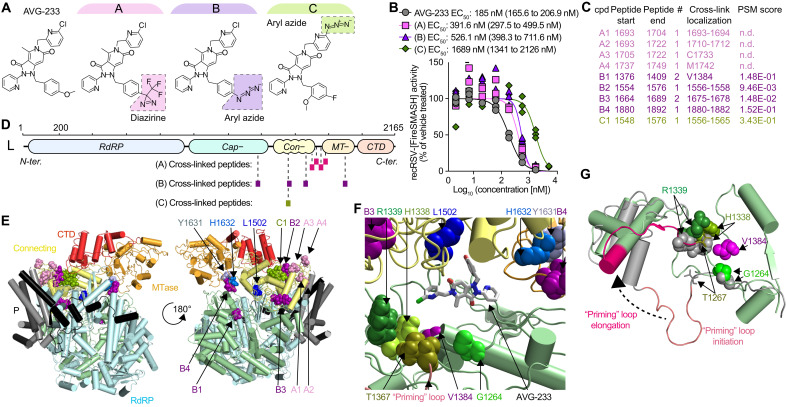
Fig. 3. AVG-233 target site mapping through photoaffinity labeling.
(A) Chemical structures of the AVG-233 analogs synthesized. Photoactivatable groups are highlighted in colored boxes. (B) Dose-response curves of the AVG-233 photoreactive analogs and standard AVG-233 against recRSV-fireSMASh. Symbols represent independent repeats (n = 3). Four-parameter variable slope regression modeling. (C) RSV L-P peptides in close proximity of bound AVG-233, identified through photoaffinity labeling and LC-MS/MS analysis. Specific residue(s) engaged (cross-link localization) and confidence [peptide-spectrum matches (PSM) score] are shown. (D) Schematic representation of the three sets of peptides identified through each photoactivatable AVG-233 analog. (E) Cartoon representation of a structural model of the RSV P-L complex in the putative pre-initiation state, based on RSV P-L reconstruction (PDB 6PZK) with residues (1461–2165) modeled after VSV P-L (PDB 6U1X). Color coding as in Fig. 1. Photocrosslinking target peptides from (C) and resistance mutations from Fig. 1 are highlighted. (F) Molecular docking of AVG-233 into proximity of photocrosslinking targets and resistance sites L1502 and H1632. Residues involved in RNA synthesis (H1338 and R1339) and the putative priming loop are highlighted. (G) Predicted priming loop positions in postulated polymerase initiation and elongation conformations. Labeled are residues identified through photocrosslinking and the adjacent, predicted priming loop pivot residue G1264.