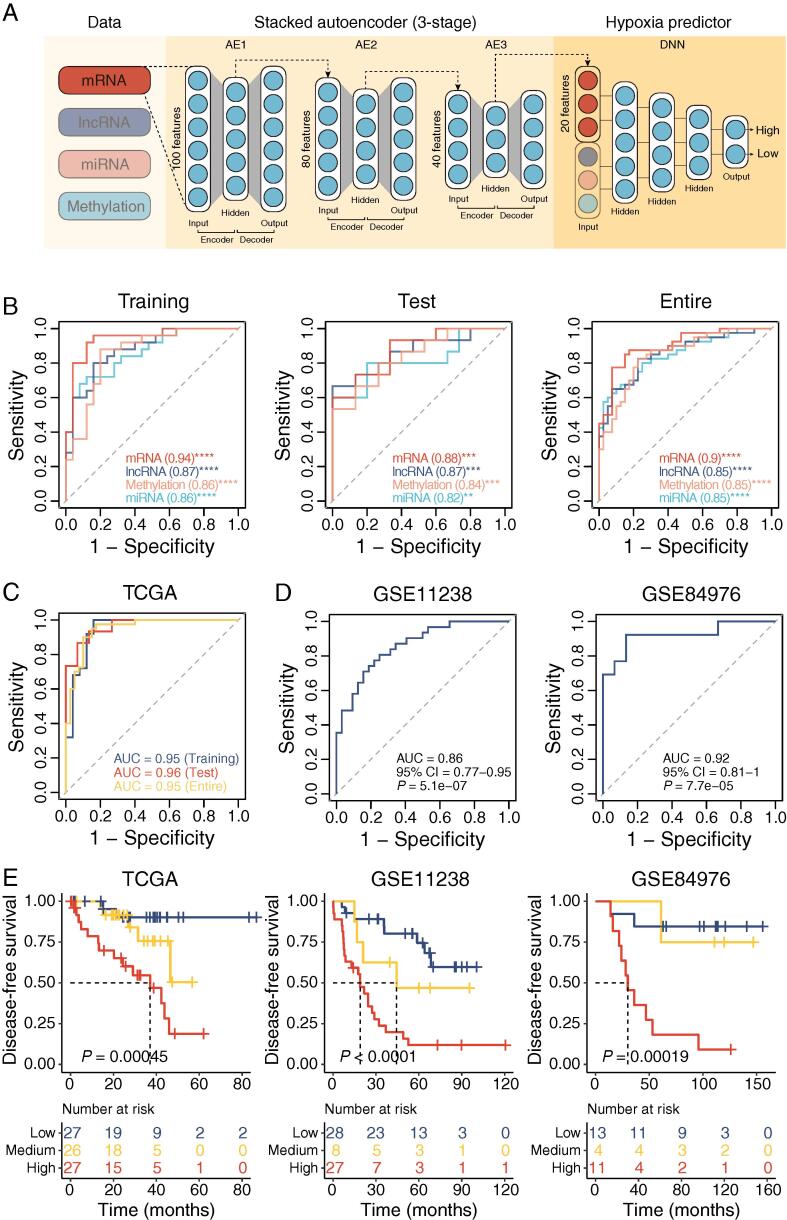
Fig. 7.
Deep neural network performance. (A) Workflow for developing deep neural network (DNN2HM) through integrating multi-omics features. TCGA cohort was split into the training cohort (50 UM tumors) and the test cohort (30 UM tumors). A three-stage stacked autoencoder was performed to learn feature structure from the training cohort and reduce the marker numbers to 20 for each molecular layer. Based on 80 UM-specific markers from four molecular layers, a deep neural network (DNN2HM) was developed to decode the hypoxic microenvironment for UM tumors. (B) Receiver-operating characteristic curves for the single molecular layer-based hypoxia predictors. Receiver-operating characteristic curves for integrated models applied on TCGA cohort (C), GSE11238 cohort, and GSE84976 cohort (D). (E) Kaplan-Meier curves of disease-free survival for high, medium, and low hypoxic tumors predicted by the DNN2HM (Log-rank test). The number of cases and events in a subgroup are shown in the plots. AE, autoencoder; DNN, deep neural network; AUC, area under the curve.