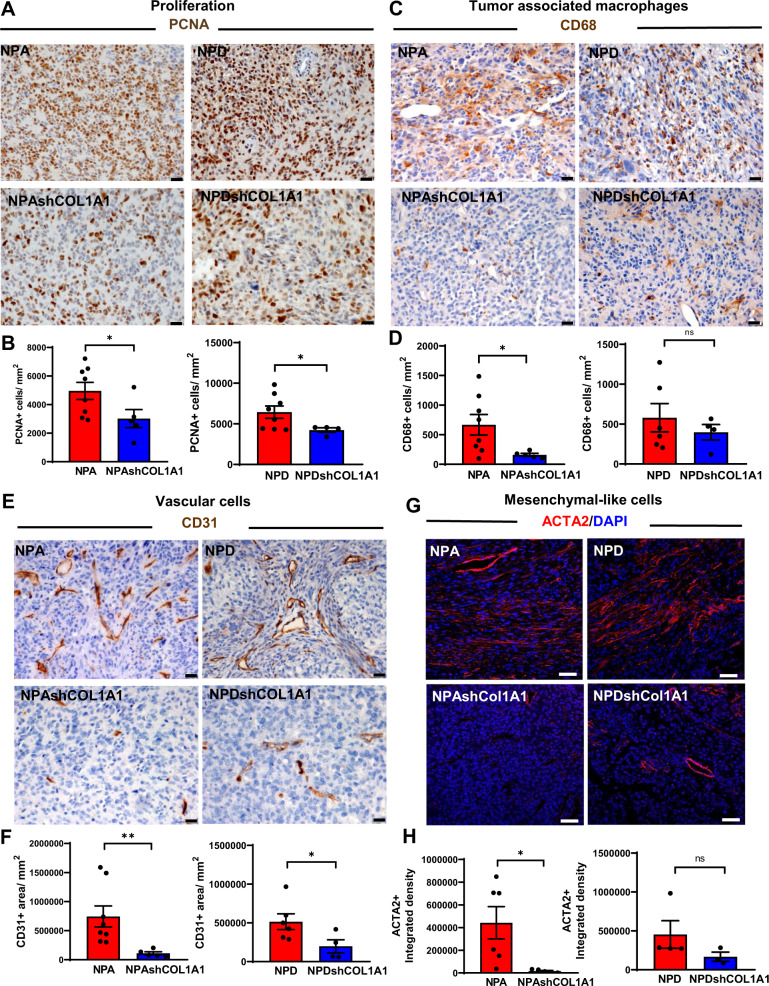
Fig. 6. Knockdown of COL1A1 within glioma cells modifies the tumor microenvironment.
Immunohistochemical analysis (A, C, and E) of GEMM of glioma controls (NPA and NPD) and COL1A1 downregulation (NPAshCOL1A1 and NPDshCOL1A1). A Representative images of PCNA expression. Scale bar: 20 μm. B Bar graphs represent the quantification of PCNA+ cells numbers (cells/mm2) using QuPath positive cell detection. Graphs present mean ± SEM, (NPA: n = 8, NPAshCOL1A1: n = 5, NPD: n = 8, NPDshCOL1A1: n = 4), two-sided t-test with Welch’s correction, *p = 0.0240. C Representative images of CD68 expression. Scale bar: 20 μm. D Bar graphs represent CD68+ cell quantification (cells/mm2) using QuPath positive cell detection. Graphs present mean ± SEM, (NPA: n = 8, NPAshCOL1A1: n = 5, NPD: n = 6, NPDshCOL1A1: n = 4), Two-sided t-test, *p < 0.05, ns: no significant. E Representative images of CD31 expression. Scale bar: 20 μm. F Bar graphs represent CD31 + cells quantification (cells/mm2) using QuPath positive cells detection. Error bars represent ± SEM, (NPA: n = 8, NPAshCOL1A1: n = 5, NPD: n = 6, NPDshCOL1A1: n = 4). Graphs present mean ± SEM. Mann–Whitney two-sided t-test, NPA vs NPAshCOL1A1 **p = 0.0016, NPD vs NPDshCOL1A1 *p = 0.0381. G Immunofluorescence analysis of GEMM of glioma controls (NPA and NPD) and COL1A1 downregulation (NPAshCOL1A1 and NPDshCOL1A1). Representative images of ACTA2 expression in red (Alexa 555) and nuclei in blue (DAPI). Scale bar: 50 μm. H Bar graphs represent ACTA2 quantification in terms of fluorescence integrated density. Graphs present mean ± SEM, (NPA: n = 6, NPAshCOL1A1: n = 5, NPD: n = 4, NPDshCOL1A1: n = 3), two-sided t-test, *p = 0.0242, ns: no significant.