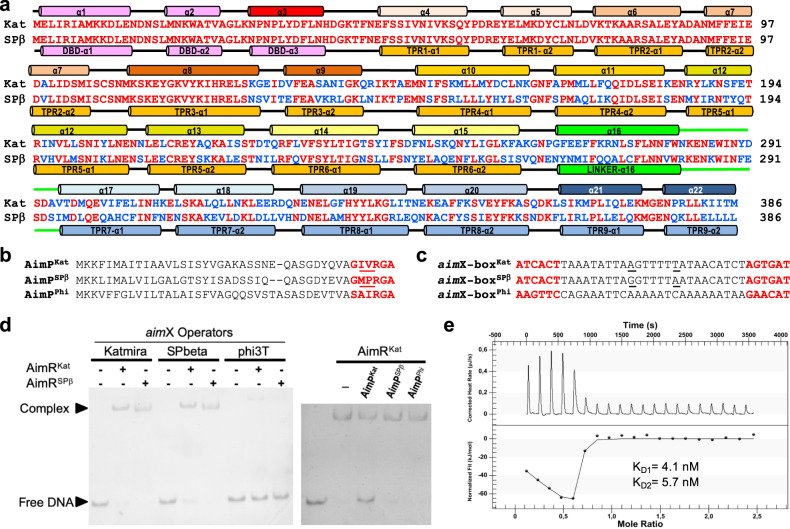
Fig. 1. AimRs present chimeric traits.
a Sequence alignment of AimRKat and AimRSPβ. Identical residues are in red colour. Structural elements are shown above and below of AimRKat and AimRSPβ sequences, respectively, and labelled by helices for the former or domains for the latter. b Sequence alignment of AimPKat, AimRSPβ and AimPPhi. Mature AimPs are highlighted in red with differential positions between AimPKat and AimRSPβ mature peptides underlined. c Sequences of DNA operators upstream of the putative aimX genes for AimRKat (aimX-boxKat), AimRSPβ (aimX-boxSPβ) and AimRPhi (aimX-boxPhi), with the 6 bp inverted repeats highlighted in red. The two different positions between aimX-boxKat and aimX-boxSPβ operator spacers are highlighted underlined. d EMSA analysis shown that both AimRKat and AimRSPβ are able to recognise reciprocally their aimX operators but not the operator for phage phi3T. The AimRKat binding to its operator is specifically disrupted by AimPKat. EMSA assays have been repeated independently three times with similar results. Source data are provided as a Source Data file. e ITC measurement of AimRKat-AimPKat binding affinity. Thermogram is adjusted to two-binding site model and the two KD (KD1 and KD2) values, one per monomer in the AimR dimer, are shown.