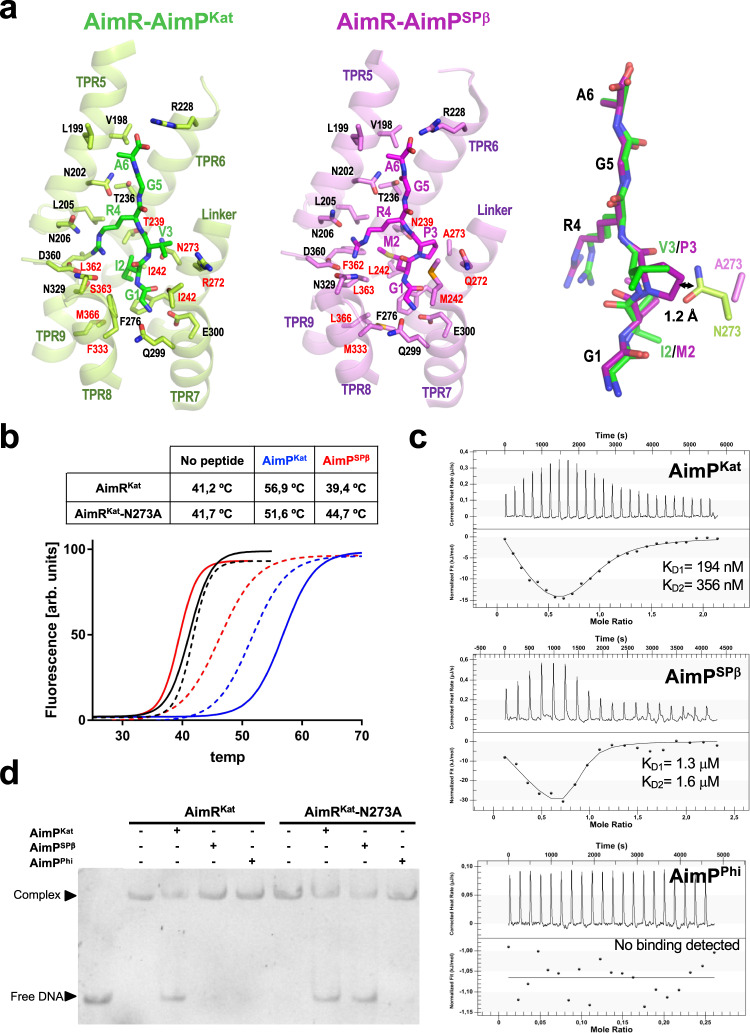
Fig. 7. AimR peptide selectivity.
a Close view of AimPs bound at the corresponding binding sites AimRKat (left) and AimRSPβ (centre) showing the AimPs and the AimR interacting residues in sticks. The AimR structural elements where the recognition residues are placed are shown in translucent cartoon and labelled. Conserved and variable interacting residues between both AimRs are labelled in black and red, respectively. In the right, the superimposition of both AimPs is shown in stick as well the side-chain for the residue in position 273. The distance between the side chains of AimRKat N273 and AimPSPβ P3 is indicated. b–d Mutation of AimRKat residue 273 alter AimP sensitivity. In (b) thermal unfolding curves of AimRKat wt (solid lines) and N273A mutant (broken lines) alone (black) or in presence of AimPKat (blue) and AimPSPβ (red) are shown. The unfolding Tms showed in the table support AimP sensitivity variation induced by the mutation. c ITC measurement of AimRKat-N273A binding affinity for AimPKat, AimPSPβ and AimPPhi. In case of binding, thermogram were adjusted to two-binding site model and the two KD (KD1 and KD2) values, one per monomer in the AimR dimer, are shown. In (d) EMSA analysis shows that both AimPKat and AimPSPβ peptides induce DNA releases for the N273A mutant form of AimRKat but only AimPKat for the wt form. Both AimRKat wt and N273A mutant are insensitive to AimPPhi. EMSA assays have been repeated independently three times with similar results. Source data are provided as a Source Data file.