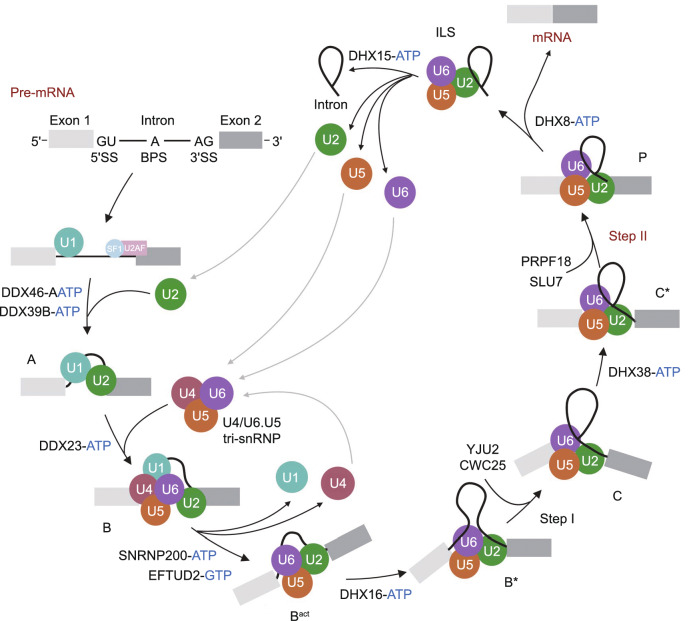
Figure 3.
Eukaryotic U2-dependent splicing cycle. The 5′SS, BPS and 3′SS are first recognized by the U1 snRNP, SF1 and U2AF, respectively, forming an early spliceosome (E complex). SF1 is displaced by the U2 snRNP to form the pre-spliceosome (A complex), which associates with the U4/U6.U5 tri-snRNP to assemble into the pre-catalytic spliceosome (B complex). B complex undergoes a series of rearrangements to form a catalytically active Bact complex and then B* complex, which carries out the first catalytic step of splicing, generating C complex. C complex undergoes additional rearrangements and then carries out the second catalytic step, resulting in a post-catalytic spliceosome (P complex) that contains the lariat intron and spliced exons. Release of the spliced exons from P complex generates the intron lariat spliceosome (ILS complex). Finally, the U2, U5 and U6 snRNPs are released from the mRNP particle and recycled for additional rounds of splicing. Each complex has a unique composition, and conversions between complexes are driven by highly conserved RNA-dependent ATPase/helicases (including DDX46, DDX39B, DDX23, SNRNP200, DHX16, DHX38, DHX8 and DHX15, and the GTPase EFTUD2)