Figure 1: Engineering circular ADAR recruiting guide RNAs (cadRNAs).
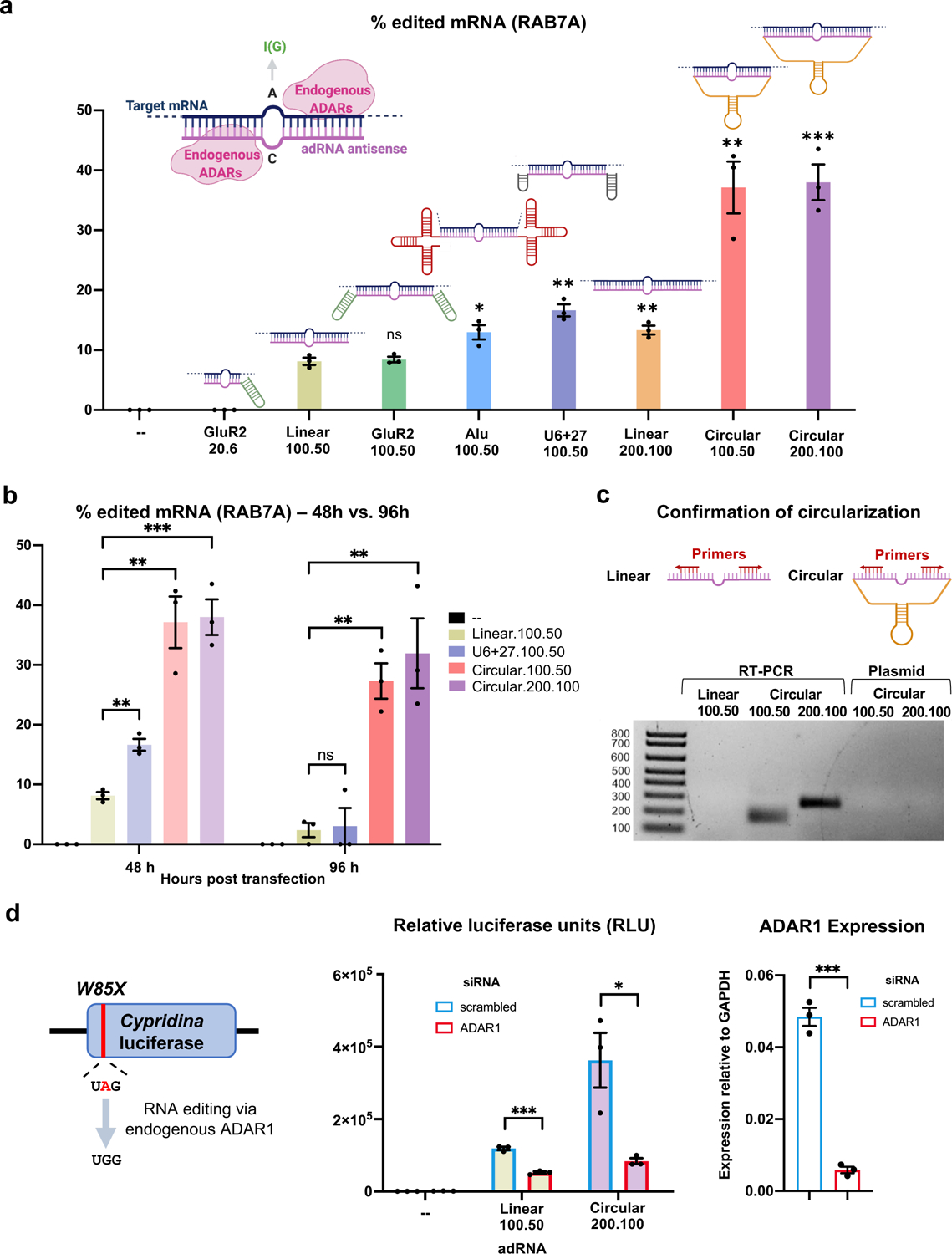
(a) A comparison of the RNA editing efficiencies in the 3’ UTR of the RAB7A transcript via various adRNA designs. Values represent mean +/− SEM (n=3; with respect to the linear.100.50, left-to-right, p=0.7289, p=0.0226, p=0.0019, p=0.0055, p=0.0027, and p=0.0006; unpaired t-test, two-tailed). In the schematics, the pink strand represents the antisense domain of the adRNA while the target mRNA is in blue. The bulge indicates the A-C mismatch between the target mRNA and adRNA. The adRNAs are labelled using the following convention: (domain name).(antisense length).(position of A-C mismatch from 5’ end of the antisense). (b) RNA editing efficiencies achieved 48 hours and 96 hours post transfection of various adRNA designs. Values represent mean +/− SEM (n=3; left-to-right, p=0.0019, p=0.0027, p=0.0006 and p=0.8488, p=0.0014, p=0.0077; unpaired t-test, two-tailed). The 48 hour panel data is reproduced from Figure 1a. (c) RT-PCR based confirmation of adRNA circularization in cells. (d) The ability of adRNAs to effect RNA editing of the cluc transcript was assessed in the presence of an siRNA targeting ADAR1. Values represent mean +/− SEM (n=3; left-to-right, p=0.0002, p=0.0216 and p=0.0001; unpaired t-test, two-tailed). All experiments were carried out in HEK293FT cells.
