Extended Data Fig. 5. Transcriptomic changes associated with in vivo cadRNA expression.
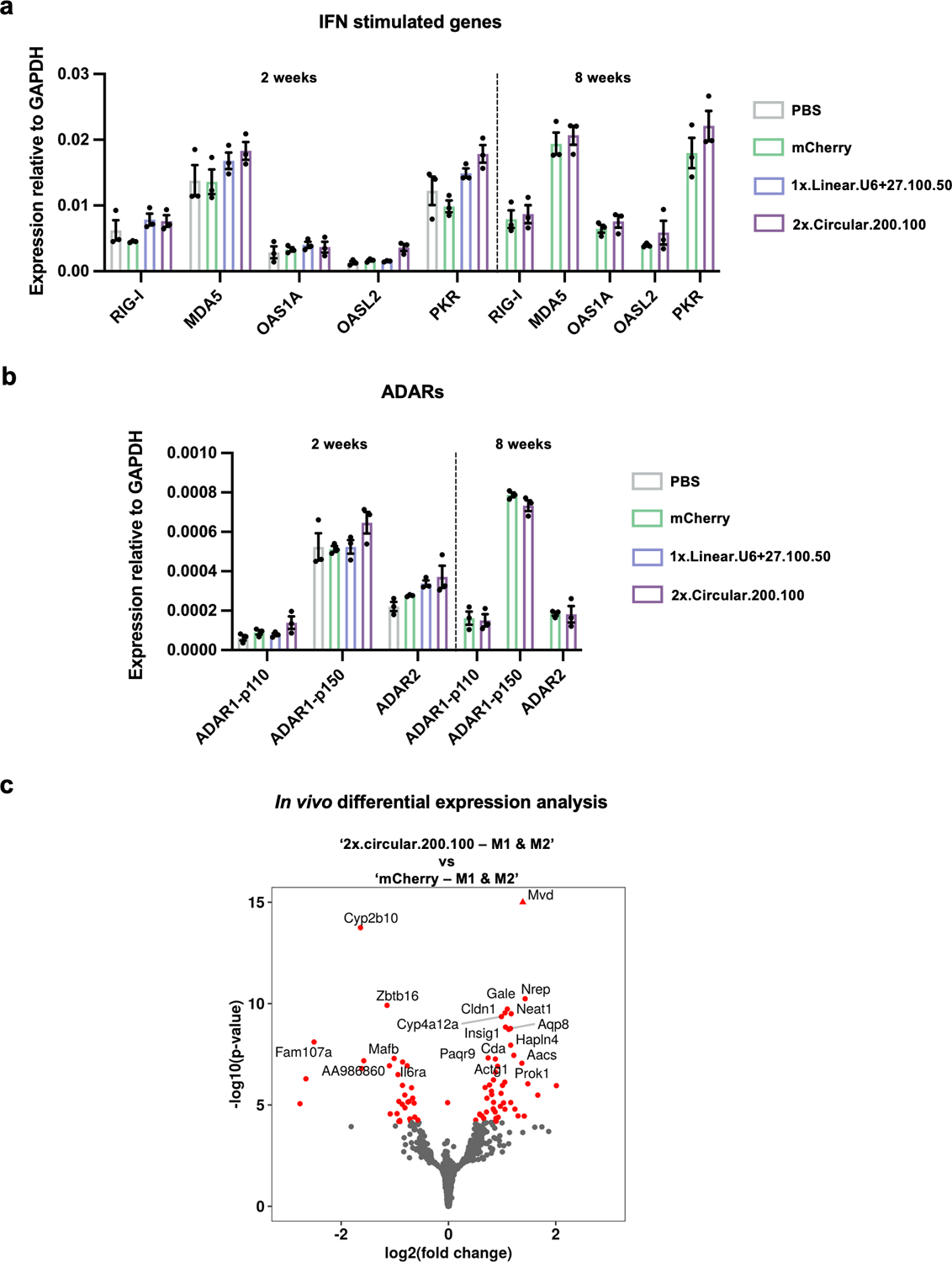
(a) qPCRs were carried out on IFN-inducible genes involved in sensing of dsRNA 2 weeks and 8 weeks post AAV injections. Values represent mean +/− SEM (n=3; p-values for 2 week long experiment, 2x.circular.200.100 vs mCherry, for genes from left to right p=0.0721, p=0.0353, p=0.8082, p=0.0748, p=0.0303; p-values for 8 week long experiment, 2x.circular.200.100 vs mCherry, for genes from left to right p=0.7276, p=0.6020, p=0.3838, p=0.3491, p=0.2746; unpaired t-test, two-tailed). (b) qPCRs were carried out on ADAR variants 2 weeks and 8 weeks post AAV injections. Values represent mean +/− SEM (n=3; p-values for 2-week long experiment, 2x.circular.200.100 vs. mCherry, for ADAR variants from left to right p=0.3165, p=0.1885, p=0.2815; p-values for 8 week long experiment, 2x.circular.200.100 vs. mCherry, for genes from left to right p=0.8150, p=0.1440, p=0.9532; unpaired t-test, two-tailed). (c) Transcriptome-wide differentially expressed genes in the two groups: 2x.circular.200.100 vs. mCherry are highlighted in red.
